class: title-slide, nobar 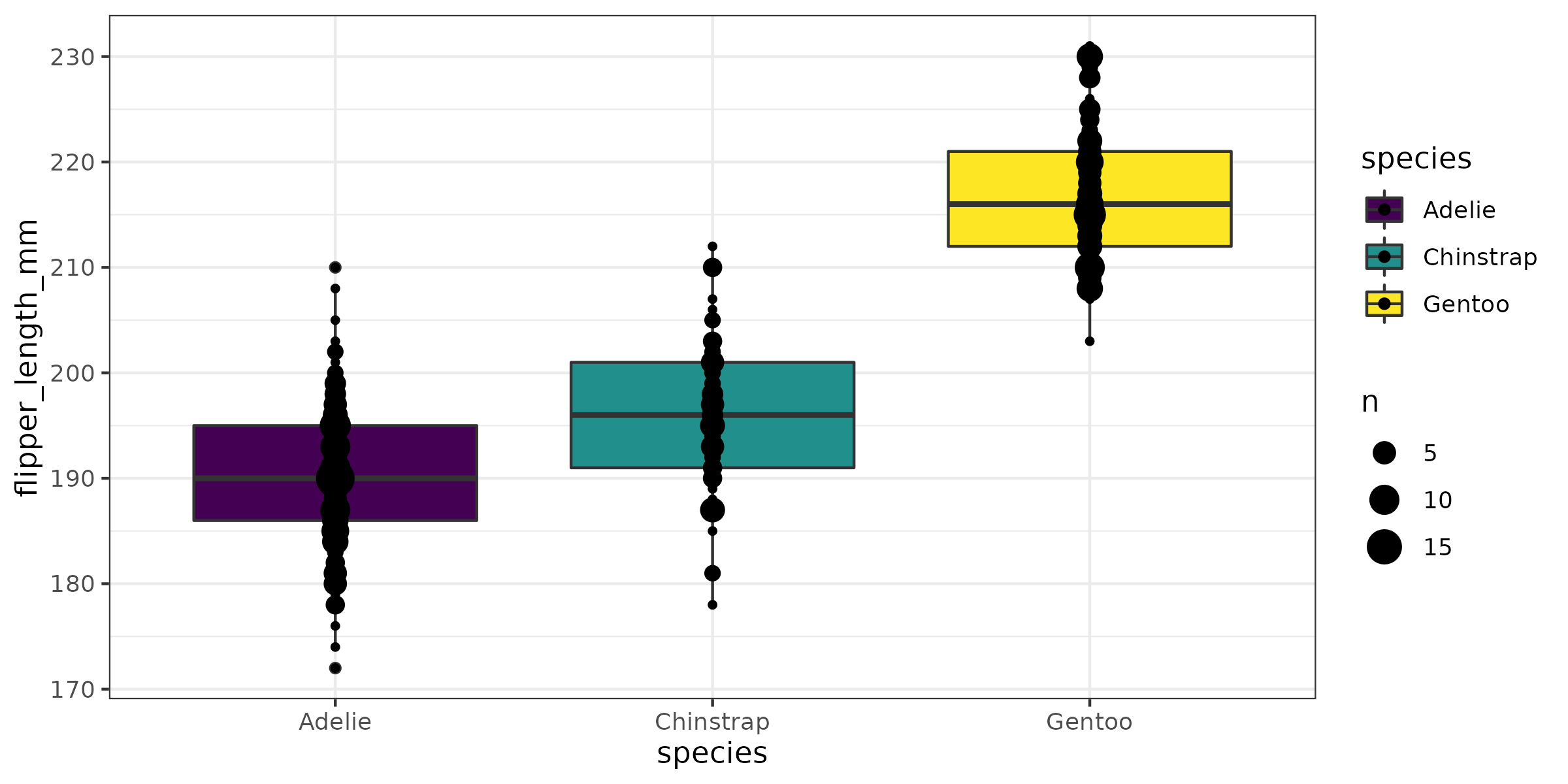 ## Workshop: Dealing with Data in R # Visualizing Data in R ## A primer on `ggplot2` .footnote[Steffi LaZerte <https://steffilazerte.ca> | *Compiled: 2022-01-26*] --- class: section # First things first Save previous script (consider names like `intro.R` or `1_getting_started.R`) Open New File <br>.medium[(make sure you're in the RStudio Project)] ![:spacer 10px]() Add `library(tidyverse)` to the top Save this new script .medium[consider names like `figs.R` or `2_figures.R`] --- # Outline ## 1. Figures with `ggplot2` .small[(A `tidyverse` package)] - Basic plot - Common plot types - Plotting by categories - Adding statistics - Customizing plots - Annotating plots ## 2. Combining figures with `patchwork` ## 3. Saving figures --- class: nobar # `ggplot2`  .footnote[Artwork by [@allison_horst](https://github.com/allisonhorst/stats-illustrations)] --- # Our data set: Palmer Penguins!  .footnote[Artwork by [@allison_horst](https://github.com/allisonhorst/stats-illustrations)]  --- # Our data set: Palmer Penguins! .small[ ```r library(palmerpenguins) penguins ``` ``` ## # A tibble: 344 × 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year ## <fct> <fct> <dbl> <dbl> <int> <int> <fct> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 male 2007 ## 2 Adelie Torgersen 39.5 17.4 186 3800 female 2007 ## 3 Adelie Torgersen 40.3 18 195 3250 female 2007 ## 4 Adelie Torgersen NA NA NA NA <NA> 2007 ## 5 Adelie Torgersen 36.7 19.3 193 3450 female 2007 ## 6 Adelie Torgersen 39.3 20.6 190 3650 male 2007 ## 7 Adelie Torgersen 38.9 17.8 181 3625 female 2007 ## 8 Adelie Torgersen 39.2 19.6 195 4675 male 2007 ## # … with 336 more rows ``` ]  .footnote[Artwork by [@allison_horst](https://github.com/allisonhorst/stats-illustrations)]  -- .medium[ > **Your turn!** > 1. Run this code and look at the output in the console > 2. Run `view(penguins)` to see the output in the built-in spreadsheet viewer ] --- # Side Note ### Where did the `penguins` data set come from? - Sometimes R packages contain data - If you load a package (i.e. `library(palmerpenguins)`) you can use the data - **Note** that here the data object is called `penguins` (not `palmerpenguins`) - **Note** this is NOT how you'll load your own data --- # A basic plot <code class ='r hljs remark-code'>library(palmerpenguins)<br>library(tidyverse)<br><br>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm)) +<br> geom_point()</code> <img src="2 Visualizing Data_files/figure-html/basic_plot-flaired-1.png" width="70%" style="display: block; margin: auto;" /> --- # Break it down <code class ='r hljs remark-code'><span style="background-color:#ffff7f">library(palmerpenguins)</span><br>library(tidyverse)<br><br>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm)) +<br> geom_point()</code> ![:spacer 10px]() ### `library(palmerpenguins)` - Load the `palmerguins` package so we have access to `penguins` data --- # Break it down <code class ='r hljs remark-code'>library(palmerpenguins)<br><span style="background-color:#ffff7f">library(tidyverse)</span><br><br>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm)) +<br> geom_point()</code> ![:spacer 10px]() ### `library(tidyverse)` - Load the `tidyverse` package (which loads the `ggplot2` package and gives us access to the `ggplot()` function among others) --- # Break it down <code class ='r hljs remark-code'>library(palmerpenguins)<br>library(tidyverse)<br><br><span style="background-color:#ffff7f">ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm))</span> +<br> geom_point()</code> ![:spacer 10px]() ### `ggplot()` - Set the attributes of your plot - **`data`** = Dataset - **`aes`** = Aesthetics (how the data are used) - Think of this as your plot defaults --- # Break it down <code class ='r hljs remark-code'>library(palmerpenguins)<br>library(tidyverse)<br><br>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm)) +<br> <span style="background-color:#ffff7f">geom_point()</span></code> ![:spacer 10px]() ### `geom_point()` - Choose a `geom` function to display the data - Always *added* to a `ggplot()` call with `+` > ggplots are essentially layered objects, starting with a call to `ggplot()` --- class: split-50 # Plots are layered .columnl[ .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-8-1.png" width="100%" style="display: block; margin: auto;" /> ]] .columnr[ .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + * geom_boxplot() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-9-1.png" width="100%" style="display: block; margin: auto;" /> ]] --- class: split-50 # Plots are layered .columnl[ .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-10-1.png" width="100%" style="display: block; margin: auto;" /> ]] .columnr[ .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + * geom_point() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-11-1.png" width="100%" style="display: block; margin: auto;" /> ]] --- class:split-50 # Plots are layered .columnl[ .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-12-1.png" width="100%" style="display: block; margin: auto;" /> ]] .columnr[ .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + * geom_violin() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-13-1.png" width="100%" style="display: block; margin: auto;" /> ]] --- class: split-50 # Plots are layered .columnl[ ### You can add multiple layers .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + geom_boxplot() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-14-1.png" width="100%" style="display: block; margin: auto;" /> ]] --- class: split-50 # Plots are layered .columnl[ ### You can add multiple layers .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + geom_boxplot() + * geom_point(size = 2, colour = "red") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-15-1.png" width="100%" style="display: block; margin: auto;" /> ]] -- .columnr[ ### Order matters .small[ ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + * geom_point(size = 2, colour = "red") + geom_boxplot() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-16-1.png" width="100%" style="display: block; margin: auto;" /> ]] --- class: split-50 # Plots are objects ### Any ggplot can be saved as an object ```r g <- ggplot(data = penguins, aes(x = sex, y = body_mass_g)) ``` -- .columnl[ ```r g ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-18-1.png" width="100%" style="display: block; margin: auto;" /> ] -- .columnr[ ```r g + geom_boxplot() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-19-1.png" width="100%" style="display: block; margin: auto;" /> ] --- class: section # More Geoms ### (Plot types) --- # Geoms: Lines ```r ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm)) + * geom_line() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-20-1.png" width="60%" style="display: block; margin: auto;" /> --- # Geoms: Boxplots ```r ggplot(data = penguins, aes(x = species, y = flipper_length_mm)) + * geom_boxplot() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-21-1.png" width="60%" style="display: block; margin: auto;" /> --- # Geoms: Histogram ```r ggplot(data = penguins, aes(x = body_mass_g)) + * geom_histogram(binwidth = 100) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-22-1.png" width="60%" style="display: block; margin: auto;" /> -- ) --- # Geoms: Barplots ### Let `ggplot` count your data ```r ggplot(data = penguins, aes(x = sex)) + * geom_bar() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-23-1.png" width="60%" style="display: block; margin: auto;" /> --- # Geoms: Barplots ### You can also provide the counts <code class ='r hljs remark-code'># Create our own data frame of counts<br>species <- count(penguins, species)<br><br>ggplot(data = species, aes(x = species, <span style="background-color:#ffff7f">y = n</span>)) +<br> geom_bar(<span style="background-color:#ffff7f">stat = "identity"</span>)</code> <img src="2 Visualizing Data_files/figure-html/bar-flaired-1.png" width="60%" style="display: block; margin: auto;" /> --- # Your Turn: Create this plot <code class ='r hljs remark-code'>library(tidyverse)<br><br>ggplot(data = <span style="background-color:#ffff7f"> </span>, aes(x = <span style="background-color:#ffff7f"> </span>, y = <span style="background-color:#ffff7f"> </span>)) +<br> geom_<span style="background-color:#ffff7f"> </span>(<span style="background-color:#ffff7f"> </span>)</code> <img src="2 Visualizing Data_files/figure-html/yt_boxplot-flaired-1.png" width="60%" style="display: block; margin: auto;" />  --- exclude: TRUE # Your Turn: Create this plot <code class ='r hljs remark-code'>library(tidyverse)<br><br>ggplot(data = penguins, aes(x = island, y = bill_depth_mm)) +<br> geom_boxplot(colour = "blue")</code> <img src="2 Visualizing Data_files/figure-html/yt_boxplot-flaired-sm1dah8-1.png" width="60%" style="display: block; margin: auto;" />  --- exclude: TRUE # Your Turn: Create this plot (Extra Challenge) <code class ='r hljs remark-code'>library(tidyverse)<br><br>ggplot(data = penguins, aes(x = island, y = bill_depth_mm)) +<br> geom_boxplot(colour = "blue") +<br> <span style="background-color:#ffff7f">geom_point()</span></code> <img src="2 Visualizing Data_files/figure-html/yt_boxplot_extra-flaired-1.png" width="60%" style="display: block; margin: auto;" />  --- exclude: TRUE # Your Turn: Create this plot (Extra Challenge) <code class ='r hljs remark-code'>library(tidyverse)<br><br>ggplot(data = penguins, aes(x = island, y = bill_depth_mm)) +<br> geom_boxplot(colour = "blue") +<br> <span style="background-color:#ffff7f">geom_count()</span></code> <img src="2 Visualizing Data_files/figure-html/yt_boxplot_extra2-flaired-1.png" width="60%" style="display: block; margin: auto;" />  --- class: section # Showing data by group --- # Mapping aesthetics ```r ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm)) + geom_point() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-29-1.png" width="60%" style="display: block; margin: auto;" /> --- # Mapping aesthetics <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, <span style="background-color:#ffff7f">colour = sex</span>)) +<br> geom_point()</code> <img src="2 Visualizing Data_files/figure-html/aes1-flaired-1.png" width="60%" style="display: block; margin: auto;" /> </code> automatically creates legends) --- # Mapping aesthetics <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, <span style="background-color:#ffff7f">size = flipper_length_mm</span>)) +<br> geom_point()</code> <img src="2 Visualizing Data_files/figure-html/aes2-flaired-1.png" width="60%" style="display: block; margin: auto;" /> --- # Mapping aesthetics <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, <span style="background-color:#ffff7f">alpha = sex</span>)) +<br> geom_point()</code> <img src="2 Visualizing Data_files/figure-html/aes3-flaired-1.png" width="60%" style="display: block; margin: auto;" /> --- # Mapping aesthetics <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, <span style="background-color:#ffff7f">shape = species</span>)) +<br> geom_point()</code> <img src="2 Visualizing Data_files/figure-html/aes4-flaired-1.png" width="60%" style="display: block; margin: auto;" /> --- # Mapping aesthetics <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, <span style="background-color:#ffff7f">colour = sex, shape = sex</span>)) +<br> geom_point()</code> <img src="2 Visualizing Data_files/figure-html/aes5-flaired-1.png" width="60%" style="display: block; margin: auto;" /> </code> combines legends where it can) --- # Faceting: `facet_wrap()` ```r ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, colour = sex)) + geom_point() + * facet_wrap(~ species) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-35-1.png" width="70%" style="display: block; margin: auto;" />  --- # Faceting: `facet_grid()` ```r ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, colour = sex)) + geom_point() + * facet_grid(sex ~ species) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-36-1.png" width="70%" style="display: block; margin: auto;" />  --- # Your Turn: Create this plot <code class ='r hljs remark-code'>ggplot(data = <span style="background-color:#ffff7f"> </span>, aes(<span style="background-color:#ffff7f"> </span>)) + <br> <span style="background-color:#ffff7f"> </span> + <br> <span style="background-color:#ffff7f"> </span></code> <img src="2 Visualizing Data_files/figure-html/yt_facet-flaired-1.png" width="80%" style="display: block; margin: auto;" /> ![:spacer 5px]() .medium[ > **Hint:** `colour` is for outlining with a colour, `fill` is for 'filling' with a colour > **Extra Challenge:** Split boxplots by sex **and** island ] --- exclude: TRUE # Your Turn: Create this plot <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = sex, y = flipper_length_mm, fill = sex)) + <br> geom_boxplot() + <br> facet_wrap(~ species)</code> <img src="2 Visualizing Data_files/figure-html/yt_facet-flaired-a9z3qju-1.png" width="80%" style="display: block; margin: auto;" /> ![:spacer 5px]() .medium[ > **Hint:** `colour` is for outlining with a colour, `fill` is for 'filling' with a colour > **Extra Challenge:** Split boxplots by sex **and** island ] --- exclude: TRUE # Your Turn: Create this plot (Extra Challenge) <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = sex, y = flipper_length_mm, <span style="background-color:#ffff7f">fill = island</span>)) + <br> geom_boxplot() + <br> facet_wrap(~ species)</code> <img src="2 Visualizing Data_files/figure-html/yt_facet_extra-flaired-1.png" width="80%" style="display: block; margin: auto;" /> ![:spacer 5px]() .medium[ > Small change (`fill = sex` to `fill = island`) results in completely different plot ] --- exclude: true class: section # Adding Statistics to Plots --- # Summarizing data ### Add data means as points ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + * stat_summary(geom = "point", fun = mean) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-40-1.png" width="60%" style="display: block; margin: auto;" /> --- # Summarizing data ### Add error bars, calculated from the data ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + stat_summary(geom = "point", fun = mean) + * stat_summary(geom = "errorbar", width = 0.05, fun.data = mean_se) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-41-1.png" width="60%" style="display: block; margin: auto;" /> --- class: section # Trendlines / Regression Lines --- # Trendlines / Regression lines ### `geom_line()` is connect-the-dots, not a trend or linear model ```r ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm)) + geom_point() + geom_line() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-42-1.png" width="60%" style="display: block; margin: auto;" /> --  --- # Trendlines / Regression lines ### Let's add a trend line properly Start with basic plot: ```r g <- ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm)) + geom_point() g ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-43-1.png" width="60%" style="display: block; margin: auto;" /> --- class: split-45 # Trendlines / Regression lines .columnl[ ### Add the `stat_smooth()` - `lm` is for "linear model" (i.e. trendline) - grey ribbon = standard error ] .columnr[ <code class ='r hljs remark-code'>g + <span style="background-color:#ffff7f">stat_smooth(method = "lm")</span></code> ] ![:spacer 55px]() <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-45-1.png" width="60%" style="display: block; margin: auto;" /> --- class: split-45 # Trendlines / Regression lines .columnl[ ### Add the `stat_smooth()` - remove the grey ribbon `se = FALSE` ] .columnr[ <code class ='r hljs remark-code'>g + stat_smooth(method = "lm", <span style="background-color:#ffff7f">se = FALSE</span>)</code> ] ![:spacer 55px]() <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-47-1.png" width="60%" style="display: block; margin: auto;" /> --- # Trendlines / Regression lines ### A line for each group - Specify group (here we use `colour` to specify `species`) ```r g <- ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, colour = species)) + geom_point() g ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-48-1.png" width="60%" style="display: block; margin: auto;" /> --- # Trendlines / Regression lines ### A line for each group - `stat_smooth()` automatically uses the same grouping ```r g + stat_smooth(method = "lm", se = FALSE) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-49-1.png" width="60%" style="display: block; margin: auto;" /> --- # Trendlines / Regression lines ### A line for each group AND overall ```r g + stat_smooth(method = "lm", se = FALSE) + stat_smooth(method = "lm", se = FALSE, colour = "black") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-50-1.png" width="60%" style="display: block; margin: auto;" /> --- # Your Turn: Create this plot - A scatter plot: **Flipper Length** by **Body Mass** grouped by **Species** - With *a single regression line for the overall trend* > **Extra Challenge:** Create a separate plot for each sex as well --- exclude: TRUE # Your Turn: Create this plot - A scatter plot: **Flipper Length** by **Body Mass** grouped by **Species** - With *a single regression line for the overall trend* ```r ggplot(data = penguins, aes(x = body_mass_g, y = flipper_length_mm, colour = species)) + geom_point() + stat_smooth(se = FALSE, colour = "black", method = "lm") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-51-1.png" width="60%" style="display: block; margin: auto;" /> --- exclude: TRUE # Your Turn: Create this plot (Extra Challenge) ```r ggplot(data = penguins, aes(x = body_mass_g, y = flipper_length_mm, colour = species)) + geom_point() + stat_smooth(se = FALSE, colour = "black", method = "lm") + * facet_wrap(~sex) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-52-1.png" width="80%" style="display: block; margin: auto;" /> --- class: section # Customizing plots --- # Customizing: Starting plot ### Let's work with this plot ```r g <- ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, colour = species)) + geom_point() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-54-1.png" width="60%" style="display: block; margin: auto;" /> --- # Customizing: Labels ```r g + labs(title = "Bill Length vs. Body Mass", x = "Body Mass (g)", y = "Bill Length (mm)", colour = "Species", tag = "A") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-55-1.png" width="60%" style="display: block; margin: auto;" /> --  --- class: split-50 # Customizing: Built-in themes .columnl[ <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-56-1.png" width="95%" style="display: block; margin: auto;" /><img src="2 Visualizing Data_files/figure-html/unnamed-chunk-56-2.png" width="95%" style="display: block; margin: auto;" /> ] .columnr[ <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-57-1.png" width="95%" style="display: block; margin: auto;" /><img src="2 Visualizing Data_files/figure-html/unnamed-chunk-57-2.png" width="95%" style="display: block; margin: auto;" /> ] --- # Customizing: Axes `scale_` + (`x` or `y`) + type (`contiuous`, `discrete`, `date`, `datetime`) - `scale_x_continuous()` - `scale_y_discrete()` - etc. ### Common arguments ```r g + scale_x_continuous(breaks = seq(0, 20, 10)) # Tick breaks g + scale_x_continuous(limits = c(0, 15)) # xlim() is a shortcut for this g + scale_x_continuous(expand = c(0, 0)) # Space between axis and data ``` --- # Customizing: Axes ### Breaks ```r g + scale_x_continuous(breaks = seq(2500, 6500, 500)) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-59-1.png" width="60%" style="display: block; margin: auto;" /> --- # Customizing: Axes ### Limits ```r g + scale_x_continuous(limits = c(3000, 4000)) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-60-1.png" width="60%" style="display: block; margin: auto;" /> --- # Customizing: Axes ### Space between origin and axis start ```r g + scale_x_continuous(expand = c(0, 0)) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-61-1.png" width="60%" style="display: block; margin: auto;" /> --- # Customizing: Aesthetics ### Using scales `scale_` + aesthetic (`colour`, `fill`, `size`, etc.) + type (`manual`, `continuous`, `datetime`, etc.) ```r g + scale_colour_manual(name = "Type", values = c("green", "purple", "yellow")) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-62-1.png" width="60%" style="display: block; margin: auto;" /> --- # Customizing: Aesthetics ### Using scales Or be very explicit: ```r g + scale_colour_manual(name = "Type", na.value = "black", values = c("Adelie" = "green", "Gentoo" = "purple", "Chinstrap" = "yellow")) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-63-1.png" width="60%" style="display: block; margin: auto;" /> --- # Customizing: Aesthetics ### For colours, consider colour-blind-friendly scale **`viridis_d` for "discrete" data** <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, colour = species)) +<br> geom_point() + <br> scale_colour<span style="background-color:#ffff7f">_viridis_d</span>(name = "Type")</code> <img src="2 Visualizing Data_files/figure-html/cb1-flaired-1.png" width="60%" style="display: block; margin: auto;" /> --- # Customizing: Aesthetics ### For colours, consider colour-blind-friendly scale **`viridis_c` for "continuous" data** <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, colour = bill_length_mm)) +<br> geom_point() + <br> scale_colour<span style="background-color:#ffff7f">_viridis_c</span>(name = "Bill Length (mm)")</code> <img src="2 Visualizing Data_files/figure-html/cb2-flaired-1.png" width="60%" style="display: block; margin: auto;" /> --- # Customizing: Aesthetics ### Forcing Remove the association between a variable and an aesthetic <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = body_mass_g, y = bill_length_mm, colour = sex)) +<br> geom_point(<span style="background-color:#ffff7f">colour = "green", size = 5</span>) +<br> stat_smooth(method = "lm", se = FALSE, <span style="background-color:#ffff7f">colour = "red"</span>)</code> <img src="2 Visualizing Data_files/figure-html/forcing-flaired-1.png" width="60%" style="display: block; margin: auto;" /> <code>) --- class: split-50 # Customizing: Legends placement .columnl[ ### At the: top, bottom, left, right ```r g + theme(legend.position = "top") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-67-1.png" width="100%" style="display: block; margin: auto;" /> ] .columnr[ ### Exactly here ```r g + theme(legend.position = c(0.15, 0.7)) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-68-1.png" width="100%" style="display: block; margin: auto;" /> ] --- # Your Turn: Create this plot ![:spacer 20px]() <img src="2 Visualizing Data_files/figure-html/yt_hard-1.png" width="90%" style="display: block; margin: auto;" />  --- exclude: TRUE # Your Turn: Create this plot .small[ ```r ggplot(penguins, aes(x = body_mass_g, y = flipper_length_mm, colour = species)) + theme_bw() + geom_point() + stat_smooth(method = "lm", se = FALSE, colour = "black") + scale_colour_viridis_d() + facet_wrap(~ sex) + labs(x = "Body Mass (g)", y = "Flipper Length (mm)", colour = "Species") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-69-1.png" width="60%" style="display: block; margin: auto;" /> ] --- class: section # Side note: Order of operations --- class: split-50 # Order of operations ## Remember... - `ggplot()` is the default line (all options passed down) - The other lines are *added* with the `+` (options only apply to this line) --- class: split-50 # Order of operations ## Where to put the `aes()`? **Sometimes it doesn't matter...** .columnl[ .small[ ```r ggplot(penguins, aes(x = body_mass_g, y = flipper_length_mm, * colour = species)) + geom_point() ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-70-1.png" width="100%" style="display: block; margin: auto;" /> ]] .columnr[ .small[ ```r ggplot(penguins, aes(x = body_mass_g, y = flipper_length_mm)) + * geom_point(aes(colour = species)) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-71-1.png" width="100%" style="display: block; margin: auto;" /> ]] --- class: split-50 # Order of operations ## Where to put the `aes()`? **Sometimes it DOES matter...** .columnl[ .small[ ```r ggplot(penguins, aes(x = body_mass_g, y = flipper_length_mm, * colour = species)) + geom_point() + stat_smooth(method = "lm") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-72-1.png" width="100%" style="display: block; margin: auto;" /> ]] .columnr[ .small[ ```r ggplot(penguins, aes(x = body_mass_g, y = flipper_length_mm)) + * geom_point(aes(colour = species)) + stat_smooth(method = "lm") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-73-1.png" width="100%" style="display: block; margin: auto;" /> ]] --  --  --- class: section # Annotating plots --- # Annotating ### Plot to be annotated: Let's add sample sizes ```r ggplot(data = penguins, aes(x = body_mass_g, y = flipper_length_mm, colour = species)) + geom_point() + facet_grid(~ species) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-74-1.png" width="100%" style="display: block; margin: auto;" /> --- # Annotating ### Create data to use in our annotations (`mutate()` is covered tomorrow!) ```r library(tidyverse) n <- count(penguins, species) n <- mutate(n, text = paste0("n = ", n)) n ``` ``` ## # A tibble: 3 × 3 ## species n text ## <fct> <int> <chr> ## 1 Adelie 152 n = 152 ## 2 Chinstrap 68 n = 68 ## 3 Gentoo 124 n = 124 ``` --- # Annotating .small[ ```r ggplot(data = penguins, aes(x = body_mass_g, y = flipper_length_mm, colour = species)) + geom_point() + facet_grid(~ species) + * geom_text(data = n, # Use 'n' data set * x = +Inf, y = -Inf, # Location relative to plot (right, bottom) * aes(label = text), # Map 'text' to label * hjust = 1, vjust = 0, # Adjust horizontal and vertical placement * colour = "black") # black text ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-76-1.png" width="100%" style="display: block; margin: auto;" /> ] --- class: section # Combining plots --- # Combining plots with `patchwork` ### Setup - Load `patchwork` - Create a couple of different plots ```r library(patchwork) g1 <- ggplot(data = penguins, aes(x = bill_length_mm, y = bill_depth_mm, colour = species)) + geom_point() g2 <- ggplot(data = penguins, aes(x = species, y = flipper_length_mm)) + geom_boxplot() g3 <- ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g, colour = species)) + geom_point() ``` --- # Combining plots with `patchwork` ### Side-by-Side 2 plots ```r g1 + g2 ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-78-1.png" width="80%" style="display: block; margin: auto;" /> --- # Combining plots with `patchwork` ### Side-by-Side 3 plots ```r g1 + g2 + g3 ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-79-1.png" width="100%" style="display: block; margin: auto;" /> --- # Combining plots with `patchwork` ### Stacked 2 plots ```r g1 / g2 ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-80-1.png" width="50%" style="display: block; margin: auto;" /> --- # Combining plots with `patchwork` ### More complex arrangements ```r g2 + (g1 / g3) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-81-1.png" width="60%" style="display: block; margin: auto;" /> --- # Combining plots with `patchwork` ### More complex arrangements ```r g2 / (g1 + g3) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-82-1.png" width="50%" style="display: block; margin: auto;" /> --- # Combining plots with `patchwork` ### "collect" common legends ```r g2 / (g1 + g3) + plot_layout(guides = "collect") ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-83-1.png" width="50%" style="display: block; margin: auto;" /> --- # Combining plots with `patchwork` ### "collect" common legends ```r g2 / (g1 + g3 + plot_layout(guides = "collect")) ``` <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-84-1.png" width="50%" style="display: block; margin: auto;" /> --- class: split-55 # Combining plots with `patchwork` .columnl[ ### Annotate .small[ ```r g2 / (g1 + g3) + plot_layout(guides = "collect") + plot_annotation(title = "Penguins Data Summary", caption = "Fig 1. Penguins Data Summary", tag_levels = "A", tag_suffix = ")") ``` ]] .columnr[ <img src="2 Visualizing Data_files/figure-html/unnamed-chunk-85-1.png" width="100%" style="display: block; margin: auto;" /> ] -- .columnl[ > **Your Turn:** Combine any 3 figures ] --- class: section # Saving plots --- # Saving plots ### RStudio Export **_Demo_** -- ### `ggsave()` ```r g <- ggplot(penguins, aes(x = sex, y = bill_length_mm, fill = year)) + geom_boxplot() ggsave(filename = "penguins_mass.png", plot = g) ``` ``` ## Saving 6 x 3.9 in image ``` --- # Saving plots ### Publication quality plots - Many publications require 'lossless' (pdf, svg, eps, ps) or high quality formats (tiff, png) - Specific sizes corresponding to columns widths - Minimum resolutions ```r g <- ggplot(penguins, aes(x = sex, y = body_mass_g)) + geom_boxplot() + labs(x = "Sex", y = "Body Mass (g)") + theme(axis.text.x = element_text(angle = 45, hjust = 1)) ggsave(filename = "penguins_mass.pdf", plot = g, dpi = 300, height = 80, width = 129, units = "mm") ``` --- # Wrapping up: Common mistakes - The **package** is `ggplot`**2**, the function is just **`ggplot()`** - Did you remember to put the **`+`** at the **end** of the line? - Order matters! If you're using custom `theme()`'s, make sure you put these lines **after** bundled themes like `theme_bw()`, or they will be overwritten - Variables like 'year' are treated as continuous, but are really categories - Wrap them in `factor()`, i.e. `ggplot(data = penguins, aes(x = factor(year), y = body_mass_g))` --- # Wrapping up: Common mistakes ### I get an error regarding an object that can't be found or aethetic length? ![:spacer 5px]() You are probably trying to plot two different datasets, and you make references to variables in the `ggplot()` call that don't exist in one of the datasets: ![:spacer 15px]() <code class ='r hljs remark-code'>n <- count(penguins, island)<br><br>ggplot(<span style="background-color:#ffff7f">data = penguins</span>, aes(x = flipper_length_mm, y = bill_length_mm, colour = species)) +<br> geom_point() +<br> facet_wrap(~ island) +<br> geom_text(<span style="background-color:#ffff7f">data = n</span>, aes(label = n), <br> x = -Inf, y = +Inf, hjust = 0, vjust = 1)</code> ``` ## Error: Aesthetics must be either length 1 or the same as the data (3): colour ``` <img src="2 Visualizing Data_files/figure-html/wu_aes1-flaired-1.png" width="60%" style="display: block; margin: auto;" /> --- # Wrapping up: Common mistakes ### I get an error regarding an object that can't be found or aethetic length? Either move the aesthetic... <code class ='r hljs remark-code'>ggplot(penguins, aes(x = flipper_length_mm, y = bill_length_mm)) + <br> geom_point(<span style="background-color:#ffff7f">aes(colour = species)</span>) +<br> facet_wrap(~ island) +<br> geom_text(data = n, aes(label = n), <br> x = -Inf, y = +Inf, hjust = 0, vjust = 1)</code> ![:spacer 15px]() Or assign it to NULL where it is missing... <code class ='r hljs remark-code'>ggplot(penguins, aes(x = flipper_length_mm, y = bill_length_mm, colour = species)) + <br> geom_point() +<br> facet_wrap(~ island) +<br> geom_text(data = n, aes(label = n, <span style="background-color:#ffff7f">colour = NULL</span>), <br> x = -Inf, y = +Inf, hjust = 0, vjust = 1)</code> --- class: space-list # Wrapping up: Further reading (all **Free**!) - RStudio > Help > Cheatsheets > Data Visualization with ggplot2 - [`ggplot2` book v3](https://ggplot2-book.org) .small[by Hadley Wickham, Danielle Navarro, and Thomas Lin Pedersen] - [R Graphics Cookbook](https://r-graphics.org/) .small[by Winston Chang] - [R for Data Science](https://r4ds.had.co.nz) .small[by Hadley Wickham and Garret Grolemund] - [Chapter on Data Visualization](http://r4ds.had.co.nz/data-visualisation.html) - [Data Visualization: A practical introduction](http://socviz.co/) .small[by Kieran Healy] - [`patchwork` website](https://patchwork.data-imaginist.com/) - [`gridExtra` Vignette: Arranging Multiple Grobs](https://cran.r-project.org/web/packages/gridExtra/vignettes/arrangeGrob.html) - Alternative to patchwork