class: title-slide, nobar  .footnote[Artwork by [@allison_horst](https://github.com/allisonhorst/stats-illustrations)] ## NRI 7350 # Data<br>Exploration .small[Also `GGally`, `skimr`, `dplyr`, and `moments`] --- # How Are we Doing? ### Take Heart! ❤️ .footnote[Artwork by [@allison_horst](https://github.com/allisonhorst/stats-illustrations)]  --- class: section # Learning to Compile Reports! Open RStudio Open your NRI project <br> Open a **old** script .small[(if it's not already open)] 'Files' pane (lower right) > Click on the script name --- 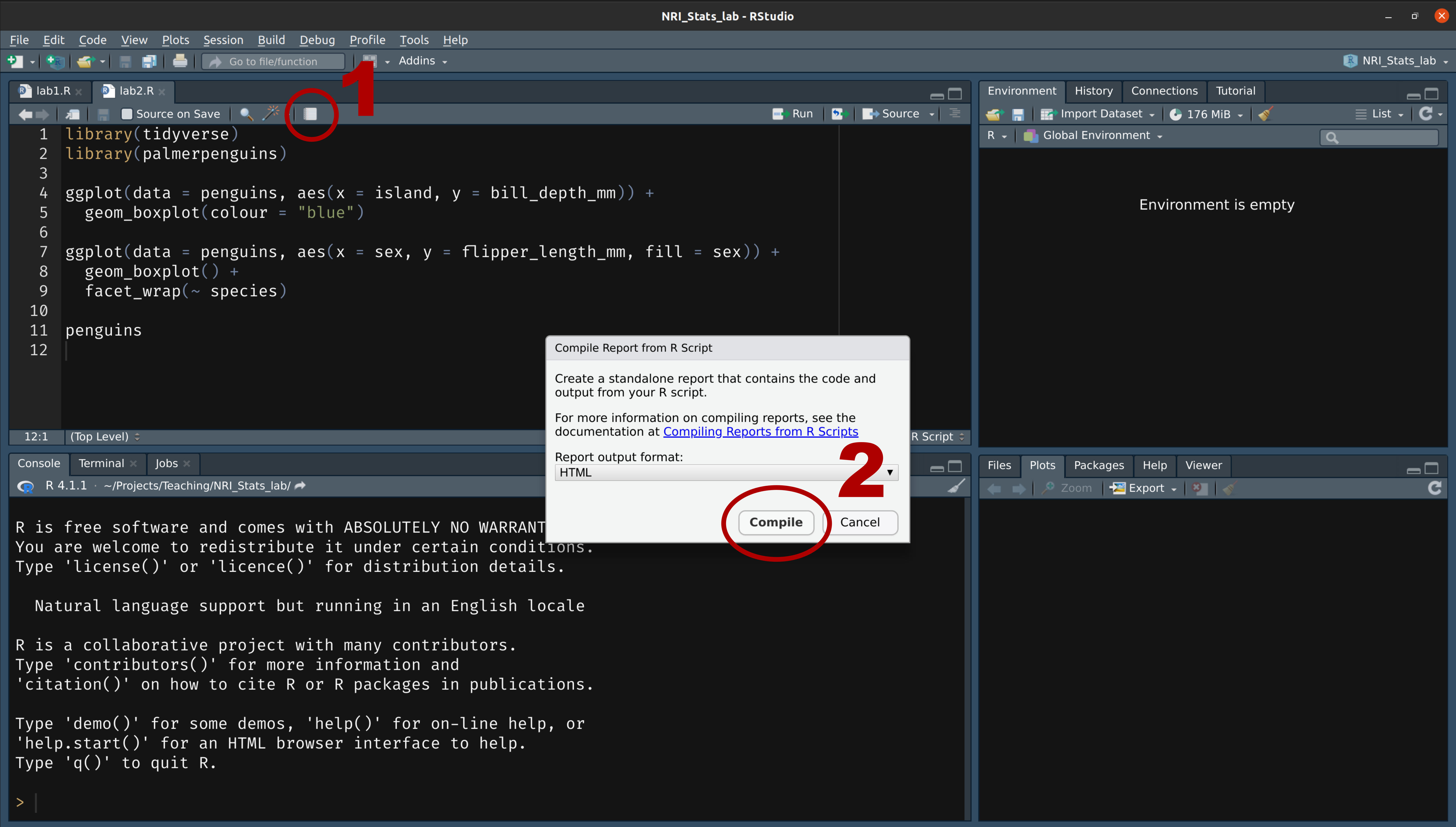 --- 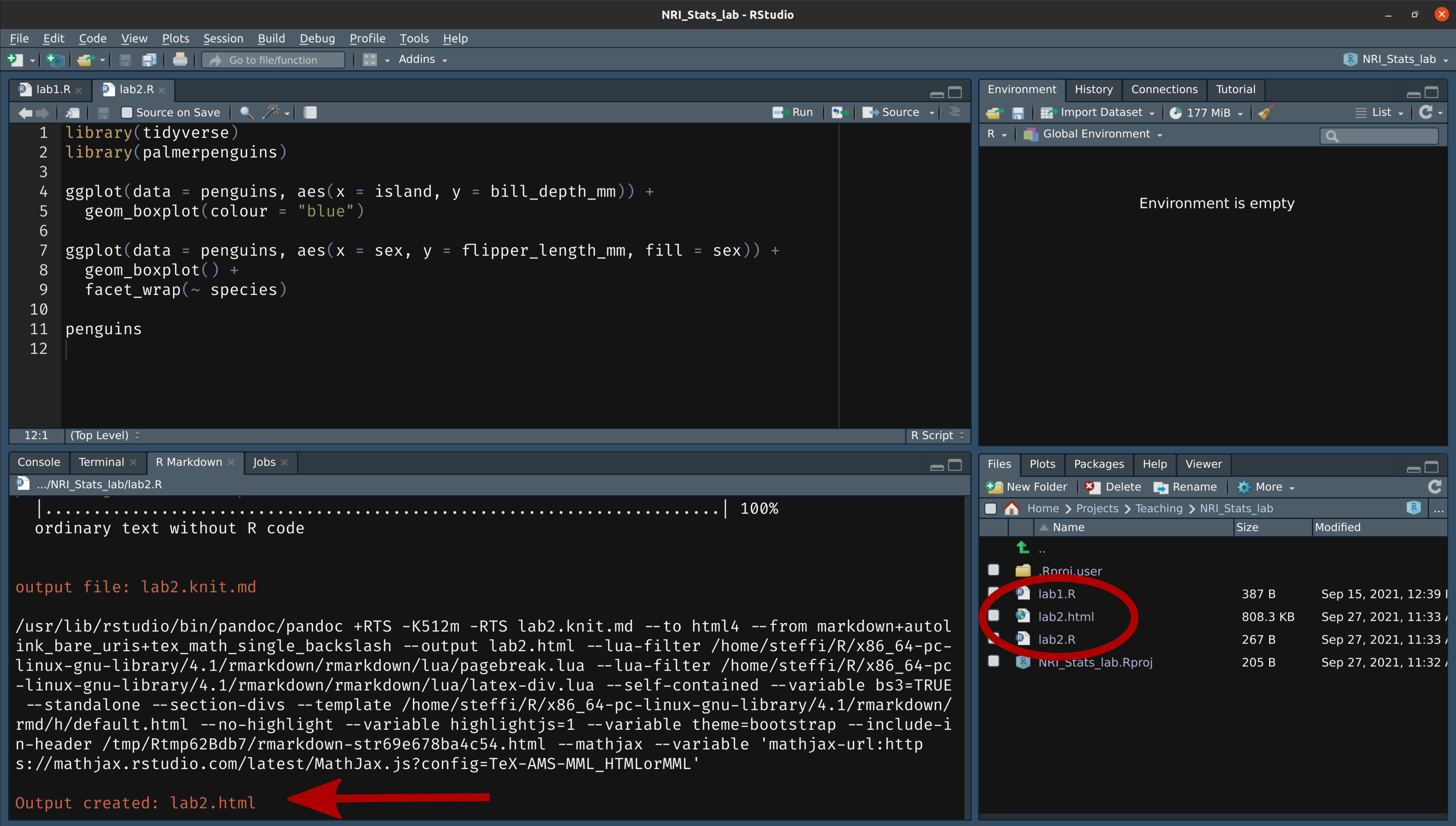 --- # Compile script into report 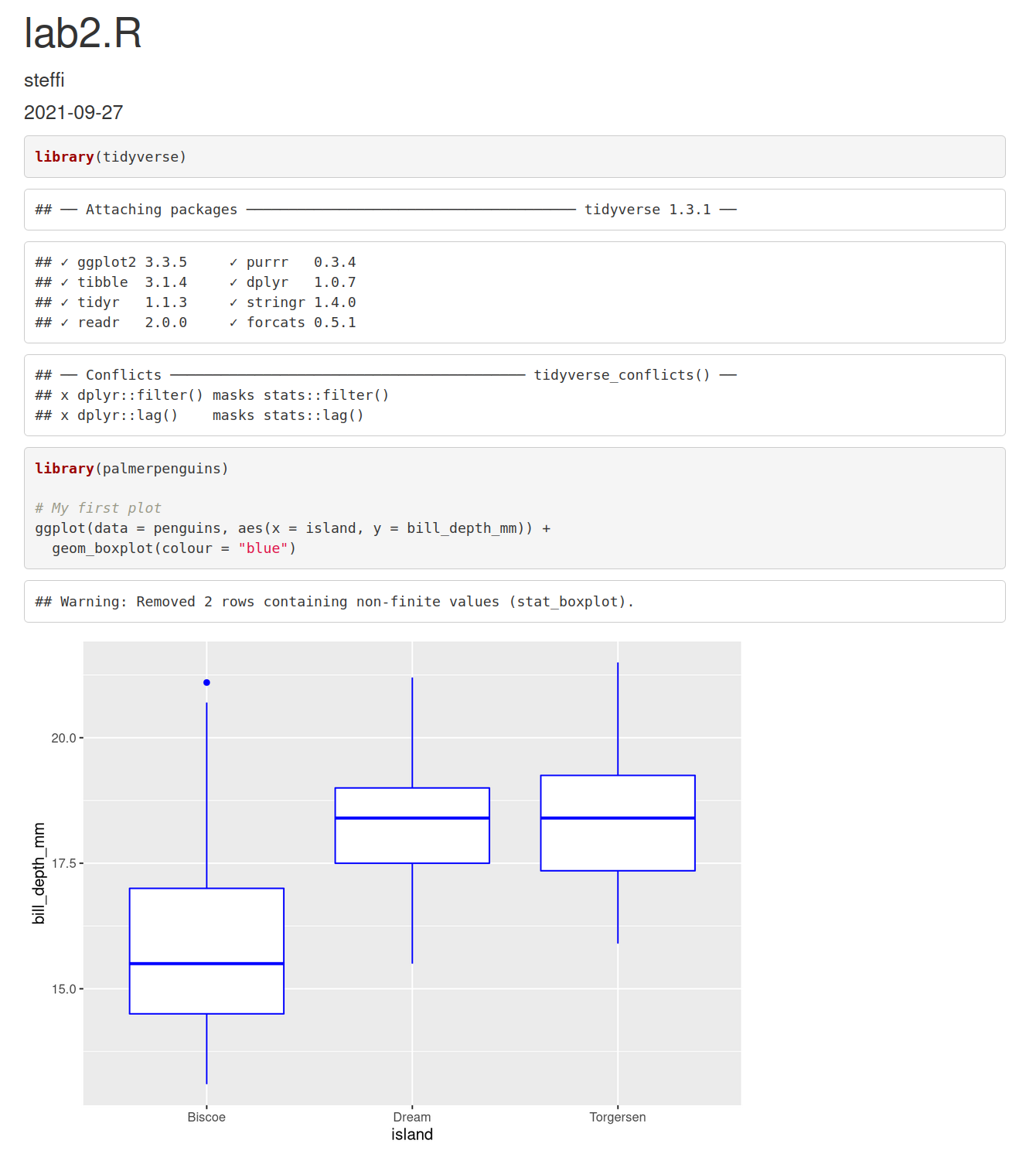 ![:spacer 100px]() > This is **all I need**: > - Your assignment answers > - This html report --- # However! ## If you want to take it further: - You can use Markdown formating to make things look nice - You just need to put `#'` in front of any Markdown notation or text (DEMO) ## Markdown - `**bold**` - `*italic*` - `#`, `##`, `###` for headings (first-, second-, third-level) ![:spacer 25px]() > **Familiar with Rmd?** > > Think of this as the mirror image: > > Instead of marking **what is** R code, you mark **what is NOT** R code > ---  --- class: split-50, space-list # Further reading  .columnl[ - [R Markdown Cookbook](https://bookdown.org/yihui/rmarkdown-cookbook/) - Chapter 3.3 [Render an R script to a report](https://bookdown.org/yihui/rmarkdown-cookbook/spin.html#spin) - [R for Data Science](https://r4ds.had.co.nz) - Chapter 27 [Rmarkdown](https://r4ds.had.co.nz/r-markdown.html) ] --- class: section # Getting started (again) **Hoping you can work with some of your own data today!** ![:spacer 15px]() Open RStudio Open your NRI project Open your data-loading script: 'Files' Pane > Click on script name **RUN YOUR SCRIPT** <br> Make sure to load packages at the top: `library(tidyverse)`<br> `library(palmerpenguins)`<br>.small[(if working with penguins today)] --- class: section # Exploring everything at once --- # Visualize with `ggpairs()` - From `GGally` package - **Caution!** If you have a lot of columns, `select()` only a few to work with <code class ='r hljs remark-code'>library(GGally)<br><br>penguins_sub <- select(penguins, -sex, -island, -year)<br>ggpairs(penguins_sub)</code> --- # Side Note: `tidyverse` functions - From `GGally` - **Caution!** If you have a lot of columns, `select()` only a few to work with <code class ='r hljs remark-code'>library(GGally)<br><br>penguins_sub <- select(<strong><span style="color:#440154">penguins,</span></strong> -<span style="color:deeppink">sex</span>, -<span style="color:deeppink">island</span>, -<span style="color:deeppink">year</span>)<br>ggpairs(penguins_sub)</code> ### `select()` - `tidyverse` functions always start with the **<span style="color:#440154">data</span>**, followed by other arguments - you can reference any **<span style="color:deeppink">column</span>** from '**<span style="color:#440154">data</span>**' - `select()` chooses columns to keep or to remove (with `-`) --- # Visualize with `ggpairs()` 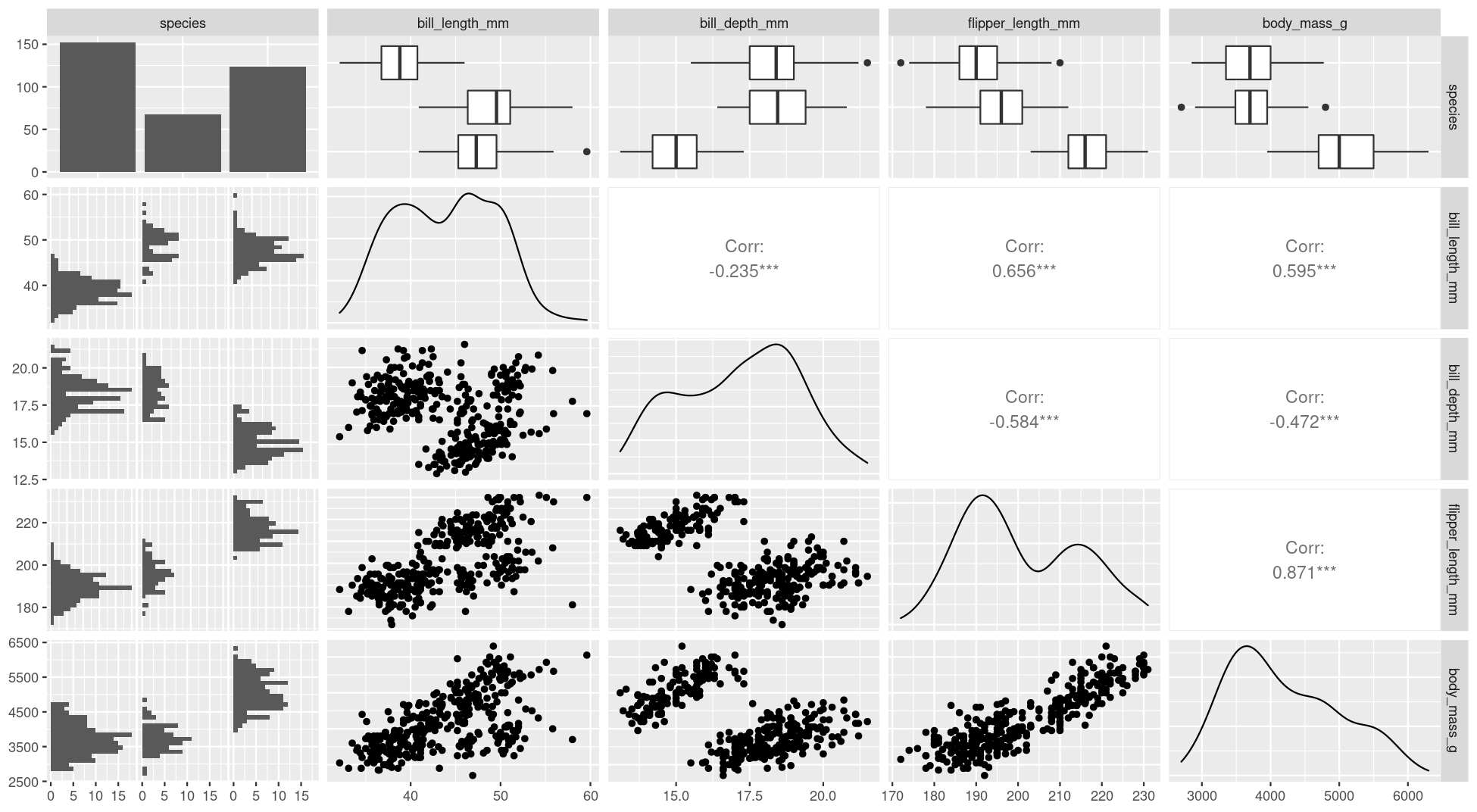 --- # Visualize with `ggpairs()` <code class ='r hljs remark-code'>library(GGally)<br><br>ggpairs(select(penguins, -sex, -island, -year), <span style="background-color:#ffff7f">aes(colour = species)</span>)</code> ![:spacer 25px]() > `ggpairs()` builds on `ggplot()` so we can use an `aes()` specification --- # Visualize with `ggpairs()` 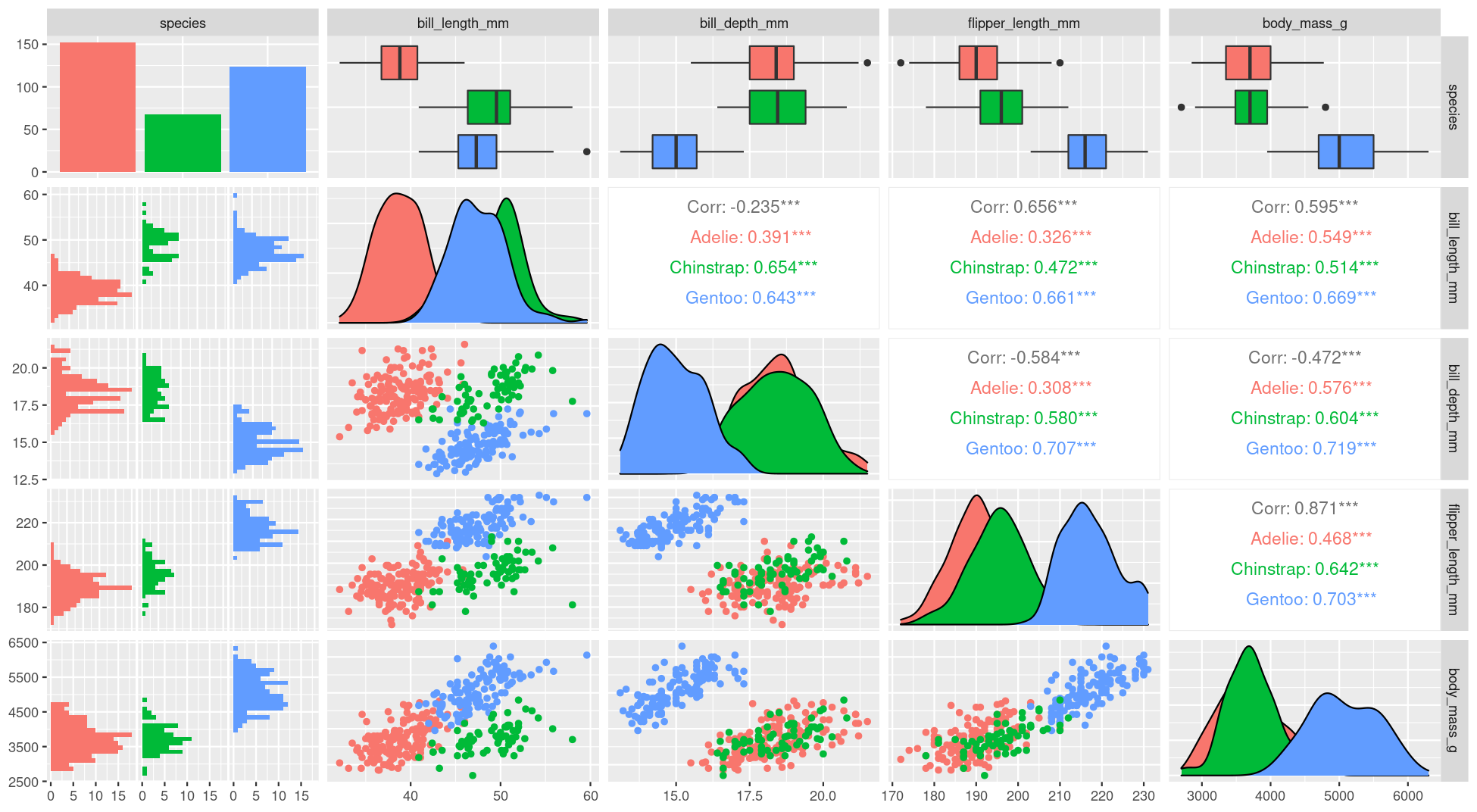 --- # Visualize with `ggpairs()` ```r library(GGally) penguins_sub <- select(penguins, -sex, -island, -year) ggpairs(penguins_sub) ggpairs(penguins_sub, aes(colour = species)) ```  --- # Summarize with `skim()` ### `skim()` from `skimr` package ```r library(skimr) skim(penguins) ``` .small[ ``` ## ── Data Summary ──────────────────────── ## Values ## Name penguins ## Number of rows 344 ## Number of columns 8 ## _______________________ ## Column type frequency: ## factor 3 ## numeric 5 ## ________________________ ## Group variables None ## ## ── Variable type: factor ─────────────────────────────────────────────────────────────────────────── ## skim_variable n_missing complete_rate ordered n_unique top_counts ## 1 species 0 1 FALSE 3 Ade: 152, Gen: 124, Chi: 68 ## 2 island 0 1 FALSE 3 Bis: 168, Dre: 124, Tor: 52 ## 3 sex 11 0.968 FALSE 2 mal: 168, fem: 165 ## ## ── Variable type: numeric ────────────────────────────────────────────────────────────────────────── ## skim_variable n_missing complete_rate mean sd p0 p25 p50 p75 p100 hist ## 1 bill_length_mm 2 0.994 43.9 5.46 32.1 39.2 44.4 48.5 59.6 ▃▇▇▆▁ ## 2 bill_depth_mm 2 0.994 17.2 1.97 13.1 15.6 17.3 18.7 21.5 ▅▅▇▇▂ ## 3 flipper_length_mm 2 0.994 201. 14.1 172 190 197 213 231 ▂▇▃▅▂ ## 4 body_mass_g 2 0.994 4202. 802. 2700 3550 4050 4750 6300 ▃▇▆▃▂ ## 5 year 0 1 2008. 0.818 2007 2007 2008 2009 2009 ▇▁▇▁▇ ``` ] --- # Summarize with `skim()` ### `skim()` from `skimr` package ```r library(skimr) skim(penguins) ``` .small[ ``` ## ## ── Variable type: factor ─────────────────────────────────────────────────────────────────────────── ## skim_variable n_missing complete_rate ordered n_unique top_counts ## 1 species 0 1 FALSE 3 Ade: 152, Gen: 124, Chi: 68 ## 2 island 0 1 FALSE 3 Bis: 168, Dre: 124, Tor: 52 ## 3 sex 11 0.968 FALSE 2 mal: 168, fem: 165 ## ## ── Variable type: numeric ────────────────────────────────────────────────────────────────────────── ## skim_variable n_missing complete_rate mean sd p0 p25 p50 p75 p100 hist ## 1 bill_length_mm 2 0.994 43.9 5.46 32.1 39.2 44.4 48.5 59.6 ▃▇▇▆▁ ## 2 bill_depth_mm 2 0.994 17.2 1.97 13.1 15.6 17.3 18.7 21.5 ▅▅▇▇▂ ## 3 flipper_length_mm 2 0.994 201. 14.1 172 190 197 213 231 ▂▇▃▅▂ ## 4 body_mass_g 2 0.994 4202. 802. 2700 3550 4050 4750 6300 ▃▇▆▃▂ ## 5 year 0 1 2008. 0.818 2007 2007 2008 2009 2009 ▇▁▇▁▇ ``` ] --  --- # Summarize with `skim()` ### `group_by` from `dplyr` package <code class ='r hljs remark-code'>penguins_sp <- group_by(penguins, species)<br>skim(penguins_sp)</code> --- # Side Note: `tidyverse` functions ### `group_by` from `dplyr` package <code class ='r hljs remark-code'>penguins_sp <- group_by(<strong><span style="color:#440154">penguins,</span></strong> <span style="color:deeppink">species</span>)<br>skim(penguins_sp)</code> ### `group_by()` - `tidyverse` functions always start with the **<span style="color:#440154">data</span>**, followed by other arguments - you can reference any **<span style="color:deeppink">column</span>** from '**<span style="color:#440154">data</span>**' - `group_by()` assigns grouping to a data frame. Here, we group `penguins` by species ![:spacer 15px]() .small[ > **Extra:** > > In the console look at `penguins` (type in `penguins` and hit enter), > and then look at `penguins_sp` (type in `penguins_sp` and it enter). > > How does the output differ? (Hint very little! But there is one difference...) ] --- # Summarize with `skim()` ### `group_by` from `dplyr` package ```r penguins_sp <- group_by(penguins, species) skim(penguins_sp) ``` .small[ ``` ## ── Data Summary ──────────────────────── ## Values ## Name skimp ## Number of rows 344 ## Number of columns 8 ## _______________________ ## Column type frequency: ## factor 2 ## numeric 5 ## ________________________ ## Group variables species ## ## ── Variable type: factor ─────────────────────────────────────────────────────────────────────────── ## skim_variable species n_missing complete_rate ordered n_unique top_counts ## 1 island Adelie 0 1 FALSE 3 Dre: 56, Tor: 52, Bis: 44 ## 2 island Chinstrap 0 1 FALSE 1 Dre: 68, Bis: 0, Tor: 0 ## 3 island Gentoo 0 1 FALSE 1 Bis: 124, Dre: 0, Tor: 0 ## 4 sex Adelie 6 0.961 FALSE 2 fem: 73, mal: 73 ## 5 sex Chinstrap 0 1 FALSE 2 fem: 34, mal: 34 ## 6 sex Gentoo 5 0.960 FALSE 2 mal: 61, fem: 58 ## ## ── Variable type: numeric ────────────────────────────────────────────────────────────────────────── ## skim_variable species n_missing complete_rate mean sd p0 p25 p50 p75 ## 1 bill_length_mm Adelie 1 0.993 38.8 2.66 32.1 36.8 38.8 40.8 ## 2 bill_length_mm Chinstrap 0 1 48.8 3.34 40.9 46.3 49.6 51.1 ## 3 bill_length_mm Gentoo 1 0.992 47.5 3.08 40.9 45.3 47.3 49.6 ## 4 bill_depth_mm Adelie 1 0.993 18.3 1.22 15.5 17.5 18.4 19 ## 5 bill_depth_mm Chinstrap 0 1 18.4 1.14 16.4 17.5 18.4 19.4 ## 6 bill_depth_mm Gentoo 1 0.992 15.0 0.981 13.1 14.2 15 15.7 ## 7 flipper_length_mm Adelie 1 0.993 190. 6.54 172 186 190 195 ## 8 flipper_length_mm Chinstrap 0 1 196. 7.13 178 191 196 201 ## 9 flipper_length_mm Gentoo 1 0.992 217. 6.48 203 212 216 221 ## 10 body_mass_g Adelie 1 0.993 3701. 459. 2850 3350 3700 4000 ## 11 body_mass_g Chinstrap 0 1 3733. 384. 2700 3488. 3700 3950 ## 12 body_mass_g Gentoo 1 0.992 5076. 504. 3950 4700 5000 5500 ## 13 year Adelie 0 1 2008. 0.822 2007 2007 2008 2009 ## 14 year Chinstrap 0 1 2008. 0.863 2007 2007 2008 2009 ## 15 year Gentoo 0 1 2008. 0.792 2007 2007 2008 2009 ## p100 hist ## 1 46 ▁▆▇▆▁ ## 2 58 ▂▇▇▅▁ ## 3 59.6 ▃▇▆▁▁ ## 4 21.5 ▂▆▇▃▁ ## 5 20.8 ▅▇▇▆▂ ## 6 17.3 ▅▇▇▆▂ ## 7 210 ▁▆▇▅▁ ## 8 212 ▁▅▇▅▂ ## 9 231 ▂▇▇▆▃ ## 10 4775 ▅▇▇▃▂ ## 11 4800 ▁▅▇▃▁ ## 12 6300 ▃▇▇▇▂ ## 13 2009 ▇▁▇▁▇ ## 14 2009 ▇▁▆▁▇ ## 15 2009 ▆▁▇▁▇ ``` ] --- # Summarize with `skim()` ### `group_by` from `dplyr` package ```r penguins_sp <- group_by(penguins, species) skim(penguins_sp) ``` .small[ ``` ## ## ── Variable type: factor ─────────────────────────────────────────────────────────────────────────── ## skim_variable species n_missing complete_rate ordered n_unique top_counts ## 1 island Adelie 0 1 FALSE 3 Dre: 56, Tor: 52, Bis: 44 ## 2 island Chinstrap 0 1 FALSE 1 Dre: 68, Bis: 0, Tor: 0 ## 3 island Gentoo 0 1 FALSE 1 Bis: 124, Dre: 0, Tor: 0 ## 4 sex Adelie 6 0.961 FALSE 2 fem: 73, mal: 73 ## 5 sex Chinstrap 0 1 FALSE 2 fem: 34, mal: 34 ## 6 sex Gentoo 5 0.960 FALSE 2 mal: 61, fem: 58 ## ## ── Variable type: numeric ────────────────────────────────────────────────────────────────────────── ## skim_variable species n_missing complete_rate mean sd p0 p25 p50 p75 ## 1 bill_length_mm Adelie 1 0.993 38.8 2.66 32.1 36.8 38.8 40.8 ## 2 bill_length_mm Chinstrap 0 1 48.8 3.34 40.9 46.3 49.6 51.1 ## 3 bill_length_mm Gentoo 1 0.992 47.5 3.08 40.9 45.3 47.3 49.6 ## 4 bill_depth_mm Adelie 1 0.993 18.3 1.22 15.5 17.5 18.4 19 ## 5 bill_depth_mm Chinstrap 0 1 18.4 1.14 16.4 17.5 18.4 19.4 ## 6 bill_depth_mm Gentoo 1 0.992 15.0 0.981 13.1 14.2 15 15.7 ## 7 flipper_length_mm Adelie 1 0.993 190. 6.54 172 186 190 195 ## 8 flipper_length_mm Chinstrap 0 1 196. 7.13 178 191 196 201 ## 9 flipper_length_mm Gentoo 1 0.992 217. 6.48 203 212 216 221 ## 10 body_mass_g Adelie 1 0.993 3701. 459. 2850 3350 3700 4000 ## 11 body_mass_g Chinstrap 0 1 3733. 384. 2700 3488. 3700 3950 ## 12 body_mass_g Gentoo 1 0.992 5076. 504. 3950 4700 5000 5500 ## 13 year Adelie 0 1 2008. 0.822 2007 2007 2008 2009 ## 14 year Chinstrap 0 1 2008. 0.863 2007 2007 2008 2009 ## 15 year Gentoo 0 1 2008. 0.792 2007 2007 2008 2009 ## p100 hist ## 1 46 ▁▆▇▆▁ ## 2 58 ▂▇▇▅▁ ## 3 59.6 ▃▇▆▁▁ ## 4 21.5 ▂▆▇▃▁ ## 5 20.8 ▅▇▇▆▂ ## 6 17.3 ▅▇▇▆▂ ## 7 210 ▁▆▇▅▁ ## 8 212 ▁▅▇▅▂ ## 9 231 ▂▇▇▆▃ ## 10 4775 ▅▇▇▃▂ ## 11 4800 ▁▅▇▃▁ ## 12 6300 ▃▇▇▇▂ ## 13 2009 ▇▁▇▁▇ ## 14 2009 ▇▁▆▁▇ ## 15 2009 ▆▁▇▁▇ ``` ] --  --- class: section # Exploring variable by variable Here, use the penguins data set (explore your own for the assignment!) --- class: split-25 # Visualize with `ggplot()` ### From last week... .columnl[ - Histograms ] .columnr[ ```r ggplot(data = penguins, aes(x = bill_length_mm)) + geom_histogram(binwidth = 0.5) ``` <img src="3 Data Exploration - answers_files/figure-html/unnamed-chunk-17-1.png" width="100%" style="display: block; margin: auto;" /> ] --- class: split-25 # Visualize with `ggplot()` ### From last week... .columnl[ - Histograms - Scatterplots ] .columnr[ ```r ggplot(data = penguins, aes(x = bill_length_mm, y = body_mass_g)) + geom_point() ``` <img src="3 Data Exploration - answers_files/figure-html/unnamed-chunk-18-1.png" width="100%" style="display: block; margin: auto;" /> ] --- class: split-25 # Visualize with `ggplot()` ### From last week... .columnl[ - Histograms - Scatterplots - Boxplots ] .columnr[ ```r ggplot(data = penguins, aes(x = species, y = body_mass_g)) + geom_boxplot() ``` <img src="3 Data Exploration - answers_files/figure-html/unnamed-chunk-19-1.png" width="100%" style="display: block; margin: auto;" /> ] --- class: split-30 # Visualize with `ggplot()` ### Histogram with Density .columnl[ - Default uses counts - Here use density `y = ..density..` - Same as density curve `geom_density()` - Use to assess shape and distribution of data ] .columnr[ <code class ='r hljs remark-code'>ggplot(data = penguins, aes(x = bill_length_mm, <span style="background-color:#ffff7f">y = ..density..</span>)) + <br> geom_histogram(binwidth = 0.5) +<br> <span style="background-color:#ffff7f">geom_density()</span></code> <img src="3 Data Exploration - answers_files/figure-html/hist-dens-flaired-1.png" width="100%" style="display: block; margin: auto;" /> ] --  --- class: split-30 # Visualize with `ggplot()` ### Histogram with Density .columnl[ - Default uses counts - Here use density `y = ..density..` - Same as density curve `geom_density()` - Use to assess shape and distribution of data ] .columnr[ ```r ggplot(data = penguins, aes(x = bill_length_mm, y = ..density.., fill = species)) + geom_histogram(binwidth = 0.5) + geom_density(alpha = 0.8) ``` <img src="3 Data Exploration - answers_files/figure-html/unnamed-chunk-21-1.png" width="100%" style="display: block; margin: auto;" /> ] --  --- class: split-30 # Visualize with `ggplot()` ### QQ Norm plots .columnl[ - Assess whether data follows normal distribution ] .columnr[ ```r ggplot(data = penguins, aes(sample = bill_length_mm)) + stat_qq() + # Add the points stat_qq_line() # Add the line ``` <img src="3 Data Exploration - answers_files/figure-html/unnamed-chunk-22-1.png" width="100%" style="display: block; margin: auto;" /> ] --  --- # Summarize with `summarize()` .small[Ha!] - From `dplyr` package (part of `tidyverse`) <code class ='r hljs remark-code'>summarize(penguins, <br> mean_mass = mean(body_mass_g),<br> sd_mass = sd(body_mass_g),<br> median_mass = median(body_mass_g))</code> --- # Side Note: `tidyverse` functions - From `dplyr` package (part of `tidyverse`) <code class ='r hljs remark-code'>summarize(<strong><span style="color:#440154">penguins</span></strong>, <br> <strong><span style="color:#277F8E">mean_mass</span></strong> = mean(<strong><span style="color:deeppink">body_mass_g</span></strong>),<br> <strong><span style="color:#277F8E">sd_mass</span></strong> = sd(<strong><span style="color:deeppink">body_mass_g</span></strong>),<br> <strong><span style="color:#277F8E">median_mass</span></strong> = median(<strong><span style="color:deeppink">body_mass_g</span></strong>))</code> ### `summarize()` - `tidyverse` functions always start with the <span style="color:#440154">**data**</span>, followed by **<span style="color:#277F8E">other arguments</span>** - you can reference any **column** from '<span style="color:#440154">**data**</span>' - `summarize()` creates a data frame with **<span style="color:#277F8E">new columns</span>** (summarizes your data) --- # Summarize with `summarize()` - From `dplyr` package (part of `tidyverse`) <code class ='r hljs remark-code'>summarize(penguins, <br> mean_mass = mean(body_mass_g),<br> sd_mass = sd(body_mass_g),<br> median_mass = median(body_mass_g))</code> ``` ## # A tibble: 1 × 3 ## mean_mass sd_mass median_mass ## <dbl> <dbl> <int> ## 1 NA NA NA ``` -- > Why all `NA`s? --- # Summarize with `summarize()` - `mean()`, `sd()`, `median()` <code class ='r hljs remark-code'>summarize(penguins, <br> mean_mass = mean(body_mass_g, <span style="background-color:#ffff7f">na.rm = TRUE</span>),<br> sd_mass = sd(body_mass_g, <span style="background-color:#ffff7f">na.rm = TRUE</span>),<br> median_mass = median(body_mass_g, <span style="background-color:#ffff7f">na.rm = TRUE</span>))</code> ``` ## # A tibble: 1 × 3 ## mean_mass sd_mass median_mass ## <dbl> <dbl> <dbl> ## 1 4202. 802. 4050 ```  --- # Summarize with `summarize()` - `mean()`, `sd()`, `median()`, `quantile()`, `n()`* <code class ='r hljs remark-code'>summarize(penguins, <br> mean_mass = mean(body_mass_g, na.rm = TRUE),<br> sd_mass = sd(body_mass_g, na.rm = TRUE),<br> median_mass = median(body_mass_g, na.rm = TRUE),<br> q25_mass = quantile(body_mass_g, probs = 0.25, na.rm = TRUE),<br> <span style="background-color:#ffff7f">n = n()</span>, # Sample size<br> <span style="background-color:#ffff7f">n_no_missing = sum(!is.na(body_mass_g)))</span> # Non-missing sample size</code> ``` ## # A tibble: 1 × 6 ## mean_mass sd_mass median_mass q25_mass n n_no_missing ## <dbl> <dbl> <dbl> <dbl> <int> <int> ## 1 4202. 802. 4050 3550 344 342 ``` .footnote[\* `n()` only works *inside* `summarize()`/`mutate()`] --- # Your Turn: `summarize()` Calculate summary statistics for **Bill Length** <code class ='r hljs remark-code'>summarize(penguins,<br> <span style="background-color:#ffff7f"> </span>bill_length_mm<span style="background-color:#ffff7f"> </span>,<br> <span style="background-color:#ffff7f"> </span>bill_length_mm<span style="background-color:#ffff7f"> </span>,<br> <span style="background-color:#ffff7f"> </span>bill_length_mm<span style="background-color:#ffff7f"> </span>,<br> <span style="background-color:#ffff7f"> </span>bill_length_mm<span style="background-color:#ffff7f"> </span><span style="background-color:#ffff7f"> </span>,<br> <span style="background-color:#ffff7f"> </span>,<br> <span style="background-color:#ffff7f"> </span>bill_length_mm<span style="background-color:#ffff7f"> </span>)</code> --- exclude: FALSE # Your Turn: `summarize()` Calculate summary statistics for **Bill Length** <code class ='r hljs remark-code'>summarize(penguins,<br> mean_bill_length = mean(bill_length_mm, na.rm = TRUE),<br> sd_bill_length = sd(bill_length_mm, na.rm = TRUE),<br> median_bill_length = median(bill_length_mm, na.rm = TRUE),<br> q25_bill_length = quantile(bill_length_mm, probs = 0.25, na.rm = TRUE),<br> n_mass = n(),<br> n_no_missing_mass = sum(!is.na(bill_length_mm)))</code> ``` ## # A tibble: 1 × 6 ## mean_bill_length sd_bill_length median_bill_length q25_bill_length n_mass n_no_missing_mass ## <dbl> <dbl> <dbl> <dbl> <int> <int> ## 1 43.9 5.46 44.4 39.2 344 342 ``` --- # Side Note: Removing `NA`s - With arguments - `na.rm = TRUE` (summary stats i.e. `mean()`, `sd()`) - `na.action = na.exclude` (models i.e., `lm()`, `lmer()`) - You can remove all `NA`s from your data (`drop_na()`) - You can selectively remove `NA`s from your data (`filter()`) --- # Side Note: Removing `NA`s ### Remove **all** `NA`s - This removes **every** row that has an `NA` in **any** column - `drop_na()` function from `tidyr` package .small[(part of `tidyverse`)] ```r penguins_no_na <- drop_na(penguins) ``` - Consider removing columns with lots of `NA`s first (assuming you don't need them) ```r *penguins_no_na <- select(penguins, -sex) penguins_no_na <- drop_na(penguins_no_na) ``` --- #Side Side Note: `tidyverse` functions - From `tidyr` package (part of `tidyverse`) <code class ='r hljs remark-code'>penguins_no_na <- drop_na<strong><span style="color:#440154">(penguins)</span></strong></code> ### `drop_na()` - `tidyverse` functions always start with the <span style="color:#440154">**data**</span>, followed by other arguments - here, there are no other arguments --- # Side Note: Removing `NA`s ### Selectively remove `NA`s with `filter()` - From `dplyr` package .small[(part of tidyverse)] ```r filter(penguins, !is.na(body_mass_g)) ``` - `is.na()` checks if there is an `NA` and returns `TRUE` if so - `!` turns a `TRUE` into a `FALSE` - `filter()` only keeps rows that are `TRUE` - **Thus** any row with an `NA` in `body_mass_g` is removed --- #Side Side Note: `tidyverse` functions - From `dplyr` package (part of `tidyverse`) <code class ='r hljs remark-code'>filter(<strong><span style="color:#440154">penguins</span></strong>, !is.na(<strong><span style="color:deeppink">body_mass_g</span></strong>))</code> ### `filter()` - `tidyverse` functions always start with the **<span style="color:#440154">data</span>**, followed by other arguments - you can reference any **<span style="color:deeppink">column</span>** from '**<span style="color:#440154">data</span>**' - `filter()` keeps only rows that return `TRUE` to the logical statements --- # Summarize with `summarize()` .small[(and `group_by()`)] - Can also use `group_by()` to calculate summaries by groups ```r *penguins_sp <- group_by(penguins, species) summarize(penguins_sp, mean_mass = mean(body_mass_g, na.rm = TRUE), sd_mass = sd(body_mass_g, na.rm = TRUE), median_mass = median(body_mass_g, na.rm = TRUE)) ``` ``` ## # A tibble: 3 × 4 ## species mean_mass sd_mass median_mass ## <fct> <dbl> <dbl> <dbl> ## 1 Adelie 3701. 459. 3700 ## 2 Chinstrap 3733. 384. 3700 ## 3 Gentoo 5076. 504. 5000 ``` --- # Summarize with `summarize()` .small[(and `group_by()`)] - Can also use `group_by()` to calculate summaries by groups ```r *penguins_sp_sex <- group_by(penguins, species, sex) summarize(penguins_sp_sex, mean_mass = mean(body_mass_g, na.rm = TRUE), sd_mass = sd(body_mass_g, na.rm = TRUE), median_mass = median(body_mass_g, na.rm = TRUE)) ``` ``` ## # A tibble: 8 × 5 ## # Groups: species [3] ## species sex mean_mass sd_mass median_mass ## <fct> <fct> <dbl> <dbl> <dbl> ## 1 Adelie female 3369. 269. 3400 ## 2 Adelie male 4043. 347. 4000 ## 3 Adelie <NA> 3540 477. 3475 ## 4 Chinstrap female 3527. 285. 3550 ## 5 Chinstrap male 3939. 362. 3950 ## 6 Gentoo female 4680. 282. 4700 ## 7 Gentoo male 5485. 313. 5500 ## 8 Gentoo <NA> 4588. 338. 4688. ``` --  --- # Side Note: Where are the decimal points? - `tibble` hides them for easy viewing ```r penguins_sum <- summarize(penguins_sp_sex, mean_mass = mean(body_mass_g, na.rm = TRUE), sd_mass = sd(body_mass_g, na.rm = TRUE), median_mass = median(body_mass_g, na.rm = TRUE)) penguins_sum ``` ``` ## # A tibble: 8 × 5 ## # Groups: species [3] ## species sex mean_mass sd_mass median_mass ## <fct> <fct> <dbl> <dbl> <dbl> ## 1 Adelie female 3369. 269. 3400 ## 2 Adelie male 4043. 347. 4000 ## 3 Adelie <NA> 3540 477. 3475 ## 4 Chinstrap female 3527. 285. 3550 ## 5 Chinstrap male 3939. 362. 3950 ## 6 Gentoo female 4680. 282. 4700 ## 7 Gentoo male 5485. 313. 5500 ## 8 Gentoo <NA> 4588. 338. 4688. ``` --  it to an object.<br>Here, <code>penguins_sum</code></small>) --- # Side Note: Where are the decimal points? - `as.data.frame()` to see the raw data ```r as.data.frame(penguins_sum) ``` ``` ## species sex mean_mass sd_mass median_mass ## 1 Adelie female 3368.836 269.3801 3400.0 ## 2 Adelie male 4043.493 346.8116 4000.0 ## 3 Adelie <NA> 3540.000 477.1661 3475.0 ## 4 Chinstrap female 3527.206 285.3339 3550.0 ## 5 Chinstrap male 3938.971 362.1376 3950.0 ## 6 Gentoo female 4679.741 281.5783 4700.0 ## 7 Gentoo male 5484.836 313.1586 5500.0 ## 8 Gentoo <NA> 4587.500 338.1937 4687.5 ``` - Or click on the name in the Environment pane --- # Side Note: Where are all my data? ```r penguins ``` ``` ## # A tibble: 344 × 8 ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## 7 Adelie Torgersen 38.9 17.8 181 3625 ## 8 Adelie Torgersen 39.2 19.6 195 4675 ## 9 Adelie Torgersen 34.1 18.1 193 3475 ## 10 Adelie Torgersen 42 20.2 190 4250 ## # … with 334 more rows, and 2 more variables: sex <fct>, year <int> ``` `... with 334 more rows, and 2 more variables: sex <fct>, year <int>` --- # Side Note: Where are all my data? ```r print(penguins, n = Inf) ``` ``` ## # A tibble: 344 × 8 ## species island bill_length_mm bill_depth_mm flipper_length_… body_mass_g ## <fct> <fct> <dbl> <dbl> <int> <int> ## 1 Adelie Torgersen 39.1 18.7 181 3750 ## 2 Adelie Torgersen 39.5 17.4 186 3800 ## 3 Adelie Torgersen 40.3 18 195 3250 ## 4 Adelie Torgersen NA NA NA NA ## 5 Adelie Torgersen 36.7 19.3 193 3450 ## 6 Adelie Torgersen 39.3 20.6 190 3650 ## 7 Adelie Torgersen 38.9 17.8 181 3625 ## 8 Adelie Torgersen 39.2 19.6 195 4675 ## 9 Adelie Torgersen 34.1 18.1 193 3475 ## 10 Adelie Torgersen 42 20.2 190 4250 ## 11 Adelie Torgersen 37.8 17.1 186 3300 ## 12 Adelie Torgersen 37.8 17.3 180 3700 ## 13 Adelie Torgersen 41.1 17.6 182 3200 ## 14 Adelie Torgersen 38.6 21.2 191 3800 ## 15 Adelie Torgersen 34.6 21.1 198 4400 ## 16 Adelie Torgersen 36.6 17.8 185 3700 ## 17 Adelie Torgersen 38.7 19 195 3450 ## 18 Adelie Torgersen 42.5 20.7 197 4500 ## 19 Adelie Torgersen 34.4 18.4 184 3325 ## 20 Adelie Torgersen 46 21.5 194 4200 ## 21 Adelie Biscoe 37.8 18.3 174 3400 ## 22 Adelie Biscoe 37.7 18.7 180 3600 ## 23 Adelie Biscoe 35.9 19.2 189 3800 ## 24 Adelie Biscoe 38.2 18.1 185 3950 ## 25 Adelie Biscoe 38.8 17.2 180 3800 ## 26 Adelie Biscoe 35.3 18.9 187 3800 ## 27 Adelie Biscoe 40.6 18.6 183 3550 ## 28 Adelie Biscoe 40.5 17.9 187 3200 ## 29 Adelie Biscoe 37.9 18.6 172 3150 ## 30 Adelie Biscoe 40.5 18.9 180 3950 ## 31 Adelie Dream 39.5 16.7 178 3250 ## 32 Adelie Dream 37.2 18.1 178 3900 ## 33 Adelie Dream 39.5 17.8 188 3300 ## 34 Adelie Dream 40.9 18.9 184 3900 ## 35 Adelie Dream 36.4 17 195 3325 ## 36 Adelie Dream 39.2 21.1 196 4150 ## 37 Adelie Dream 38.8 20 190 3950 ## 38 Adelie Dream 42.2 18.5 180 3550 ## 39 Adelie Dream 37.6 19.3 181 3300 ## 40 Adelie Dream 39.8 19.1 184 4650 ## 41 Adelie Dream 36.5 18 182 3150 ## 42 Adelie Dream 40.8 18.4 195 3900 ## 43 Adelie Dream 36 18.5 186 3100 ## 44 Adelie Dream 44.1 19.7 196 4400 ## 45 Adelie Dream 37 16.9 185 3000 ## 46 Adelie Dream 39.6 18.8 190 4600 ## 47 Adelie Dream 41.1 19 182 3425 ## 48 Adelie Dream 37.5 18.9 179 2975 ## 49 Adelie Dream 36 17.9 190 3450 ## 50 Adelie Dream 42.3 21.2 191 4150 ## 51 Adelie Biscoe 39.6 17.7 186 3500 ## 52 Adelie Biscoe 40.1 18.9 188 4300 ## 53 Adelie Biscoe 35 17.9 190 3450 ## 54 Adelie Biscoe 42 19.5 200 4050 ## 55 Adelie Biscoe 34.5 18.1 187 2900 ## 56 Adelie Biscoe 41.4 18.6 191 3700 ## 57 Adelie Biscoe 39 17.5 186 3550 ## 58 Adelie Biscoe 40.6 18.8 193 3800 ## 59 Adelie Biscoe 36.5 16.6 181 2850 ## 60 Adelie Biscoe 37.6 19.1 194 3750 ## 61 Adelie Biscoe 35.7 16.9 185 3150 ## 62 Adelie Biscoe 41.3 21.1 195 4400 ## 63 Adelie Biscoe 37.6 17 185 3600 ## 64 Adelie Biscoe 41.1 18.2 192 4050 ## 65 Adelie Biscoe 36.4 17.1 184 2850 ## 66 Adelie Biscoe 41.6 18 192 3950 ## 67 Adelie Biscoe 35.5 16.2 195 3350 ## 68 Adelie Biscoe 41.1 19.1 188 4100 ## 69 Adelie Torgersen 35.9 16.6 190 3050 ## 70 Adelie Torgersen 41.8 19.4 198 4450 ## 71 Adelie Torgersen 33.5 19 190 3600 ## 72 Adelie Torgersen 39.7 18.4 190 3900 ## 73 Adelie Torgersen 39.6 17.2 196 3550 ## 74 Adelie Torgersen 45.8 18.9 197 4150 ## 75 Adelie Torgersen 35.5 17.5 190 3700 ## 76 Adelie Torgersen 42.8 18.5 195 4250 ## 77 Adelie Torgersen 40.9 16.8 191 3700 ## 78 Adelie Torgersen 37.2 19.4 184 3900 ## 79 Adelie Torgersen 36.2 16.1 187 3550 ## 80 Adelie Torgersen 42.1 19.1 195 4000 ## 81 Adelie Torgersen 34.6 17.2 189 3200 ## 82 Adelie Torgersen 42.9 17.6 196 4700 ## 83 Adelie Torgersen 36.7 18.8 187 3800 ## 84 Adelie Torgersen 35.1 19.4 193 4200 ## 85 Adelie Dream 37.3 17.8 191 3350 ## 86 Adelie Dream 41.3 20.3 194 3550 ## 87 Adelie Dream 36.3 19.5 190 3800 ## 88 Adelie Dream 36.9 18.6 189 3500 ## 89 Adelie Dream 38.3 19.2 189 3950 ## 90 Adelie Dream 38.9 18.8 190 3600 ## 91 Adelie Dream 35.7 18 202 3550 ## 92 Adelie Dream 41.1 18.1 205 4300 ## 93 Adelie Dream 34 17.1 185 3400 ## 94 Adelie Dream 39.6 18.1 186 4450 ## 95 Adelie Dream 36.2 17.3 187 3300 ## 96 Adelie Dream 40.8 18.9 208 4300 ## 97 Adelie Dream 38.1 18.6 190 3700 ## 98 Adelie Dream 40.3 18.5 196 4350 ## 99 Adelie Dream 33.1 16.1 178 2900 ## 100 Adelie Dream 43.2 18.5 192 4100 ## 101 Adelie Biscoe 35 17.9 192 3725 ## 102 Adelie Biscoe 41 20 203 4725 ## 103 Adelie Biscoe 37.7 16 183 3075 ## 104 Adelie Biscoe 37.8 20 190 4250 ## 105 Adelie Biscoe 37.9 18.6 193 2925 ## 106 Adelie Biscoe 39.7 18.9 184 3550 ## 107 Adelie Biscoe 38.6 17.2 199 3750 ## 108 Adelie Biscoe 38.2 20 190 3900 ## 109 Adelie Biscoe 38.1 17 181 3175 ## 110 Adelie Biscoe 43.2 19 197 4775 ## 111 Adelie Biscoe 38.1 16.5 198 3825 ## 112 Adelie Biscoe 45.6 20.3 191 4600 ## 113 Adelie Biscoe 39.7 17.7 193 3200 ## 114 Adelie Biscoe 42.2 19.5 197 4275 ## 115 Adelie Biscoe 39.6 20.7 191 3900 ## 116 Adelie Biscoe 42.7 18.3 196 4075 ## 117 Adelie Torgersen 38.6 17 188 2900 ## 118 Adelie Torgersen 37.3 20.5 199 3775 ## 119 Adelie Torgersen 35.7 17 189 3350 ## 120 Adelie Torgersen 41.1 18.6 189 3325 ## 121 Adelie Torgersen 36.2 17.2 187 3150 ## 122 Adelie Torgersen 37.7 19.8 198 3500 ## 123 Adelie Torgersen 40.2 17 176 3450 ## 124 Adelie Torgersen 41.4 18.5 202 3875 ## 125 Adelie Torgersen 35.2 15.9 186 3050 ## 126 Adelie Torgersen 40.6 19 199 4000 ## 127 Adelie Torgersen 38.8 17.6 191 3275 ## 128 Adelie Torgersen 41.5 18.3 195 4300 ## 129 Adelie Torgersen 39 17.1 191 3050 ## 130 Adelie Torgersen 44.1 18 210 4000 ## 131 Adelie Torgersen 38.5 17.9 190 3325 ## 132 Adelie Torgersen 43.1 19.2 197 3500 ## 133 Adelie Dream 36.8 18.5 193 3500 ## 134 Adelie Dream 37.5 18.5 199 4475 ## 135 Adelie Dream 38.1 17.6 187 3425 ## 136 Adelie Dream 41.1 17.5 190 3900 ## 137 Adelie Dream 35.6 17.5 191 3175 ## 138 Adelie Dream 40.2 20.1 200 3975 ## 139 Adelie Dream 37 16.5 185 3400 ## 140 Adelie Dream 39.7 17.9 193 4250 ## 141 Adelie Dream 40.2 17.1 193 3400 ## 142 Adelie Dream 40.6 17.2 187 3475 ## 143 Adelie Dream 32.1 15.5 188 3050 ## 144 Adelie Dream 40.7 17 190 3725 ## 145 Adelie Dream 37.3 16.8 192 3000 ## 146 Adelie Dream 39 18.7 185 3650 ## 147 Adelie Dream 39.2 18.6 190 4250 ## 148 Adelie Dream 36.6 18.4 184 3475 ## 149 Adelie Dream 36 17.8 195 3450 ## 150 Adelie Dream 37.8 18.1 193 3750 ## 151 Adelie Dream 36 17.1 187 3700 ## 152 Adelie Dream 41.5 18.5 201 4000 ## 153 Gentoo Biscoe 46.1 13.2 211 4500 ## 154 Gentoo Biscoe 50 16.3 230 5700 ## 155 Gentoo Biscoe 48.7 14.1 210 4450 ## 156 Gentoo Biscoe 50 15.2 218 5700 ## 157 Gentoo Biscoe 47.6 14.5 215 5400 ## 158 Gentoo Biscoe 46.5 13.5 210 4550 ## 159 Gentoo Biscoe 45.4 14.6 211 4800 ## 160 Gentoo Biscoe 46.7 15.3 219 5200 ## 161 Gentoo Biscoe 43.3 13.4 209 4400 ## 162 Gentoo Biscoe 46.8 15.4 215 5150 ## 163 Gentoo Biscoe 40.9 13.7 214 4650 ## 164 Gentoo Biscoe 49 16.1 216 5550 ## 165 Gentoo Biscoe 45.5 13.7 214 4650 ## 166 Gentoo Biscoe 48.4 14.6 213 5850 ## 167 Gentoo Biscoe 45.8 14.6 210 4200 ## 168 Gentoo Biscoe 49.3 15.7 217 5850 ## 169 Gentoo Biscoe 42 13.5 210 4150 ## 170 Gentoo Biscoe 49.2 15.2 221 6300 ## 171 Gentoo Biscoe 46.2 14.5 209 4800 ## 172 Gentoo Biscoe 48.7 15.1 222 5350 ## 173 Gentoo Biscoe 50.2 14.3 218 5700 ## 174 Gentoo Biscoe 45.1 14.5 215 5000 ## 175 Gentoo Biscoe 46.5 14.5 213 4400 ## 176 Gentoo Biscoe 46.3 15.8 215 5050 ## 177 Gentoo Biscoe 42.9 13.1 215 5000 ## 178 Gentoo Biscoe 46.1 15.1 215 5100 ## 179 Gentoo Biscoe 44.5 14.3 216 4100 ## 180 Gentoo Biscoe 47.8 15 215 5650 ## 181 Gentoo Biscoe 48.2 14.3 210 4600 ## 182 Gentoo Biscoe 50 15.3 220 5550 ## 183 Gentoo Biscoe 47.3 15.3 222 5250 ## 184 Gentoo Biscoe 42.8 14.2 209 4700 ## 185 Gentoo Biscoe 45.1 14.5 207 5050 ## 186 Gentoo Biscoe 59.6 17 230 6050 ## 187 Gentoo Biscoe 49.1 14.8 220 5150 ## 188 Gentoo Biscoe 48.4 16.3 220 5400 ## 189 Gentoo Biscoe 42.6 13.7 213 4950 ## 190 Gentoo Biscoe 44.4 17.3 219 5250 ## 191 Gentoo Biscoe 44 13.6 208 4350 ## 192 Gentoo Biscoe 48.7 15.7 208 5350 ## 193 Gentoo Biscoe 42.7 13.7 208 3950 ## 194 Gentoo Biscoe 49.6 16 225 5700 ## 195 Gentoo Biscoe 45.3 13.7 210 4300 ## 196 Gentoo Biscoe 49.6 15 216 4750 ## 197 Gentoo Biscoe 50.5 15.9 222 5550 ## 198 Gentoo Biscoe 43.6 13.9 217 4900 ## 199 Gentoo Biscoe 45.5 13.9 210 4200 ## 200 Gentoo Biscoe 50.5 15.9 225 5400 ## 201 Gentoo Biscoe 44.9 13.3 213 5100 ## 202 Gentoo Biscoe 45.2 15.8 215 5300 ## 203 Gentoo Biscoe 46.6 14.2 210 4850 ## 204 Gentoo Biscoe 48.5 14.1 220 5300 ## 205 Gentoo Biscoe 45.1 14.4 210 4400 ## 206 Gentoo Biscoe 50.1 15 225 5000 ## 207 Gentoo Biscoe 46.5 14.4 217 4900 ## 208 Gentoo Biscoe 45 15.4 220 5050 ## 209 Gentoo Biscoe 43.8 13.9 208 4300 ## 210 Gentoo Biscoe 45.5 15 220 5000 ## 211 Gentoo Biscoe 43.2 14.5 208 4450 ## 212 Gentoo Biscoe 50.4 15.3 224 5550 ## 213 Gentoo Biscoe 45.3 13.8 208 4200 ## 214 Gentoo Biscoe 46.2 14.9 221 5300 ## 215 Gentoo Biscoe 45.7 13.9 214 4400 ## 216 Gentoo Biscoe 54.3 15.7 231 5650 ## 217 Gentoo Biscoe 45.8 14.2 219 4700 ## 218 Gentoo Biscoe 49.8 16.8 230 5700 ## 219 Gentoo Biscoe 46.2 14.4 214 4650 ## 220 Gentoo Biscoe 49.5 16.2 229 5800 ## 221 Gentoo Biscoe 43.5 14.2 220 4700 ## 222 Gentoo Biscoe 50.7 15 223 5550 ## 223 Gentoo Biscoe 47.7 15 216 4750 ## 224 Gentoo Biscoe 46.4 15.6 221 5000 ## 225 Gentoo Biscoe 48.2 15.6 221 5100 ## 226 Gentoo Biscoe 46.5 14.8 217 5200 ## 227 Gentoo Biscoe 46.4 15 216 4700 ## 228 Gentoo Biscoe 48.6 16 230 5800 ## 229 Gentoo Biscoe 47.5 14.2 209 4600 ## 230 Gentoo Biscoe 51.1 16.3 220 6000 ## 231 Gentoo Biscoe 45.2 13.8 215 4750 ## 232 Gentoo Biscoe 45.2 16.4 223 5950 ## 233 Gentoo Biscoe 49.1 14.5 212 4625 ## 234 Gentoo Biscoe 52.5 15.6 221 5450 ## 235 Gentoo Biscoe 47.4 14.6 212 4725 ## 236 Gentoo Biscoe 50 15.9 224 5350 ## 237 Gentoo Biscoe 44.9 13.8 212 4750 ## 238 Gentoo Biscoe 50.8 17.3 228 5600 ## 239 Gentoo Biscoe 43.4 14.4 218 4600 ## 240 Gentoo Biscoe 51.3 14.2 218 5300 ## 241 Gentoo Biscoe 47.5 14 212 4875 ## 242 Gentoo Biscoe 52.1 17 230 5550 ## 243 Gentoo Biscoe 47.5 15 218 4950 ## 244 Gentoo Biscoe 52.2 17.1 228 5400 ## 245 Gentoo Biscoe 45.5 14.5 212 4750 ## 246 Gentoo Biscoe 49.5 16.1 224 5650 ## 247 Gentoo Biscoe 44.5 14.7 214 4850 ## 248 Gentoo Biscoe 50.8 15.7 226 5200 ## 249 Gentoo Biscoe 49.4 15.8 216 4925 ## 250 Gentoo Biscoe 46.9 14.6 222 4875 ## 251 Gentoo Biscoe 48.4 14.4 203 4625 ## 252 Gentoo Biscoe 51.1 16.5 225 5250 ## 253 Gentoo Biscoe 48.5 15 219 4850 ## 254 Gentoo Biscoe 55.9 17 228 5600 ## 255 Gentoo Biscoe 47.2 15.5 215 4975 ## 256 Gentoo Biscoe 49.1 15 228 5500 ## 257 Gentoo Biscoe 47.3 13.8 216 4725 ## 258 Gentoo Biscoe 46.8 16.1 215 5500 ## 259 Gentoo Biscoe 41.7 14.7 210 4700 ## 260 Gentoo Biscoe 53.4 15.8 219 5500 ## 261 Gentoo Biscoe 43.3 14 208 4575 ## 262 Gentoo Biscoe 48.1 15.1 209 5500 ## 263 Gentoo Biscoe 50.5 15.2 216 5000 ## 264 Gentoo Biscoe 49.8 15.9 229 5950 ## 265 Gentoo Biscoe 43.5 15.2 213 4650 ## 266 Gentoo Biscoe 51.5 16.3 230 5500 ## 267 Gentoo Biscoe 46.2 14.1 217 4375 ## 268 Gentoo Biscoe 55.1 16 230 5850 ## 269 Gentoo Biscoe 44.5 15.7 217 4875 ## 270 Gentoo Biscoe 48.8 16.2 222 6000 ## 271 Gentoo Biscoe 47.2 13.7 214 4925 ## 272 Gentoo Biscoe NA NA NA NA ## 273 Gentoo Biscoe 46.8 14.3 215 4850 ## 274 Gentoo Biscoe 50.4 15.7 222 5750 ## 275 Gentoo Biscoe 45.2 14.8 212 5200 ## 276 Gentoo Biscoe 49.9 16.1 213 5400 ## 277 Chinstrap Dream 46.5 17.9 192 3500 ## 278 Chinstrap Dream 50 19.5 196 3900 ## 279 Chinstrap Dream 51.3 19.2 193 3650 ## 280 Chinstrap Dream 45.4 18.7 188 3525 ## 281 Chinstrap Dream 52.7 19.8 197 3725 ## 282 Chinstrap Dream 45.2 17.8 198 3950 ## 283 Chinstrap Dream 46.1 18.2 178 3250 ## 284 Chinstrap Dream 51.3 18.2 197 3750 ## 285 Chinstrap Dream 46 18.9 195 4150 ## 286 Chinstrap Dream 51.3 19.9 198 3700 ## 287 Chinstrap Dream 46.6 17.8 193 3800 ## 288 Chinstrap Dream 51.7 20.3 194 3775 ## 289 Chinstrap Dream 47 17.3 185 3700 ## 290 Chinstrap Dream 52 18.1 201 4050 ## 291 Chinstrap Dream 45.9 17.1 190 3575 ## 292 Chinstrap Dream 50.5 19.6 201 4050 ## 293 Chinstrap Dream 50.3 20 197 3300 ## 294 Chinstrap Dream 58 17.8 181 3700 ## 295 Chinstrap Dream 46.4 18.6 190 3450 ## 296 Chinstrap Dream 49.2 18.2 195 4400 ## 297 Chinstrap Dream 42.4 17.3 181 3600 ## 298 Chinstrap Dream 48.5 17.5 191 3400 ## 299 Chinstrap Dream 43.2 16.6 187 2900 ## 300 Chinstrap Dream 50.6 19.4 193 3800 ## 301 Chinstrap Dream 46.7 17.9 195 3300 ## 302 Chinstrap Dream 52 19 197 4150 ## 303 Chinstrap Dream 50.5 18.4 200 3400 ## 304 Chinstrap Dream 49.5 19 200 3800 ## 305 Chinstrap Dream 46.4 17.8 191 3700 ## 306 Chinstrap Dream 52.8 20 205 4550 ## 307 Chinstrap Dream 40.9 16.6 187 3200 ## 308 Chinstrap Dream 54.2 20.8 201 4300 ## 309 Chinstrap Dream 42.5 16.7 187 3350 ## 310 Chinstrap Dream 51 18.8 203 4100 ## 311 Chinstrap Dream 49.7 18.6 195 3600 ## 312 Chinstrap Dream 47.5 16.8 199 3900 ## 313 Chinstrap Dream 47.6 18.3 195 3850 ## 314 Chinstrap Dream 52 20.7 210 4800 ## 315 Chinstrap Dream 46.9 16.6 192 2700 ## 316 Chinstrap Dream 53.5 19.9 205 4500 ## 317 Chinstrap Dream 49 19.5 210 3950 ## 318 Chinstrap Dream 46.2 17.5 187 3650 ## 319 Chinstrap Dream 50.9 19.1 196 3550 ## 320 Chinstrap Dream 45.5 17 196 3500 ## 321 Chinstrap Dream 50.9 17.9 196 3675 ## 322 Chinstrap Dream 50.8 18.5 201 4450 ## 323 Chinstrap Dream 50.1 17.9 190 3400 ## 324 Chinstrap Dream 49 19.6 212 4300 ## 325 Chinstrap Dream 51.5 18.7 187 3250 ## 326 Chinstrap Dream 49.8 17.3 198 3675 ## 327 Chinstrap Dream 48.1 16.4 199 3325 ## 328 Chinstrap Dream 51.4 19 201 3950 ## 329 Chinstrap Dream 45.7 17.3 193 3600 ## 330 Chinstrap Dream 50.7 19.7 203 4050 ## 331 Chinstrap Dream 42.5 17.3 187 3350 ## 332 Chinstrap Dream 52.2 18.8 197 3450 ## 333 Chinstrap Dream 45.2 16.6 191 3250 ## 334 Chinstrap Dream 49.3 19.9 203 4050 ## 335 Chinstrap Dream 50.2 18.8 202 3800 ## 336 Chinstrap Dream 45.6 19.4 194 3525 ## 337 Chinstrap Dream 51.9 19.5 206 3950 ## 338 Chinstrap Dream 46.8 16.5 189 3650 ## 339 Chinstrap Dream 45.7 17 195 3650 ## 340 Chinstrap Dream 55.8 19.8 207 4000 ## 341 Chinstrap Dream 43.5 18.1 202 3400 ## 342 Chinstrap Dream 49.6 18.2 193 3775 ## 343 Chinstrap Dream 50.8 19 210 4100 ## 344 Chinstrap Dream 50.2 18.7 198 3775 ## # … with 2 more variables: sex <fct>, year <int> ``` --- # Side Note: Where are all my data? ```r as.data.frame(penguins) ``` ``` ## species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g sex year ## 1 Adelie Torgersen 39.1 18.7 181 3750 male 2007 ## 2 Adelie Torgersen 39.5 17.4 186 3800 female 2007 ## 3 Adelie Torgersen 40.3 18.0 195 3250 female 2007 ## 4 Adelie Torgersen NA NA NA NA <NA> 2007 ## 5 Adelie Torgersen 36.7 19.3 193 3450 female 2007 ## 6 Adelie Torgersen 39.3 20.6 190 3650 male 2007 ## 7 Adelie Torgersen 38.9 17.8 181 3625 female 2007 ## 8 Adelie Torgersen 39.2 19.6 195 4675 male 2007 ## 9 Adelie Torgersen 34.1 18.1 193 3475 <NA> 2007 ## 10 Adelie Torgersen 42.0 20.2 190 4250 <NA> 2007 ## 11 Adelie Torgersen 37.8 17.1 186 3300 <NA> 2007 ## 12 Adelie Torgersen 37.8 17.3 180 3700 <NA> 2007 ## 13 Adelie Torgersen 41.1 17.6 182 3200 female 2007 ## 14 Adelie Torgersen 38.6 21.2 191 3800 male 2007 ## 15 Adelie Torgersen 34.6 21.1 198 4400 male 2007 ## 16 Adelie Torgersen 36.6 17.8 185 3700 female 2007 ## 17 Adelie Torgersen 38.7 19.0 195 3450 female 2007 ## 18 Adelie Torgersen 42.5 20.7 197 4500 male 2007 ## 19 Adelie Torgersen 34.4 18.4 184 3325 female 2007 ## 20 Adelie Torgersen 46.0 21.5 194 4200 male 2007 ## 21 Adelie Biscoe 37.8 18.3 174 3400 female 2007 ## 22 Adelie Biscoe 37.7 18.7 180 3600 male 2007 ## 23 Adelie Biscoe 35.9 19.2 189 3800 female 2007 ## 24 Adelie Biscoe 38.2 18.1 185 3950 male 2007 ## 25 Adelie Biscoe 38.8 17.2 180 3800 male 2007 ## 26 Adelie Biscoe 35.3 18.9 187 3800 female 2007 ## 27 Adelie Biscoe 40.6 18.6 183 3550 male 2007 ## 28 Adelie Biscoe 40.5 17.9 187 3200 female 2007 ## 29 Adelie Biscoe 37.9 18.6 172 3150 female 2007 ## 30 Adelie Biscoe 40.5 18.9 180 3950 male 2007 ## 31 Adelie Dream 39.5 16.7 178 3250 female 2007 ## 32 Adelie Dream 37.2 18.1 178 3900 male 2007 ## 33 Adelie Dream 39.5 17.8 188 3300 female 2007 ## 34 Adelie Dream 40.9 18.9 184 3900 male 2007 ## 35 Adelie Dream 36.4 17.0 195 3325 female 2007 ## 36 Adelie Dream 39.2 21.1 196 4150 male 2007 ## 37 Adelie Dream 38.8 20.0 190 3950 male 2007 ## 38 Adelie Dream 42.2 18.5 180 3550 female 2007 ## 39 Adelie Dream 37.6 19.3 181 3300 female 2007 ## 40 Adelie Dream 39.8 19.1 184 4650 male 2007 ## 41 Adelie Dream 36.5 18.0 182 3150 female 2007 ## 42 Adelie Dream 40.8 18.4 195 3900 male 2007 ## 43 Adelie Dream 36.0 18.5 186 3100 female 2007 ## 44 Adelie Dream 44.1 19.7 196 4400 male 2007 ## 45 Adelie Dream 37.0 16.9 185 3000 female 2007 ## 46 Adelie Dream 39.6 18.8 190 4600 male 2007 ## 47 Adelie Dream 41.1 19.0 182 3425 male 2007 ## 48 Adelie Dream 37.5 18.9 179 2975 <NA> 2007 ## 49 Adelie Dream 36.0 17.9 190 3450 female 2007 ## 50 Adelie Dream 42.3 21.2 191 4150 male 2007 ## 51 Adelie Biscoe 39.6 17.7 186 3500 female 2008 ## 52 Adelie Biscoe 40.1 18.9 188 4300 male 2008 ## 53 Adelie Biscoe 35.0 17.9 190 3450 female 2008 ## 54 Adelie Biscoe 42.0 19.5 200 4050 male 2008 ## 55 Adelie Biscoe 34.5 18.1 187 2900 female 2008 ## 56 Adelie Biscoe 41.4 18.6 191 3700 male 2008 ## 57 Adelie Biscoe 39.0 17.5 186 3550 female 2008 ## 58 Adelie Biscoe 40.6 18.8 193 3800 male 2008 ## 59 Adelie Biscoe 36.5 16.6 181 2850 female 2008 ## 60 Adelie Biscoe 37.6 19.1 194 3750 male 2008 ## 61 Adelie Biscoe 35.7 16.9 185 3150 female 2008 ## 62 Adelie Biscoe 41.3 21.1 195 4400 male 2008 ## 63 Adelie Biscoe 37.6 17.0 185 3600 female 2008 ## 64 Adelie Biscoe 41.1 18.2 192 4050 male 2008 ## 65 Adelie Biscoe 36.4 17.1 184 2850 female 2008 ## 66 Adelie Biscoe 41.6 18.0 192 3950 male 2008 ## 67 Adelie Biscoe 35.5 16.2 195 3350 female 2008 ## 68 Adelie Biscoe 41.1 19.1 188 4100 male 2008 ## 69 Adelie Torgersen 35.9 16.6 190 3050 female 2008 ## 70 Adelie Torgersen 41.8 19.4 198 4450 male 2008 ## 71 Adelie Torgersen 33.5 19.0 190 3600 female 2008 ## 72 Adelie Torgersen 39.7 18.4 190 3900 male 2008 ## 73 Adelie Torgersen 39.6 17.2 196 3550 female 2008 ## 74 Adelie Torgersen 45.8 18.9 197 4150 male 2008 ## 75 Adelie Torgersen 35.5 17.5 190 3700 female 2008 ## 76 Adelie Torgersen 42.8 18.5 195 4250 male 2008 ## 77 Adelie Torgersen 40.9 16.8 191 3700 female 2008 ## 78 Adelie Torgersen 37.2 19.4 184 3900 male 2008 ## 79 Adelie Torgersen 36.2 16.1 187 3550 female 2008 ## 80 Adelie Torgersen 42.1 19.1 195 4000 male 2008 ## 81 Adelie Torgersen 34.6 17.2 189 3200 female 2008 ## 82 Adelie Torgersen 42.9 17.6 196 4700 male 2008 ## 83 Adelie Torgersen 36.7 18.8 187 3800 female 2008 ## 84 Adelie Torgersen 35.1 19.4 193 4200 male 2008 ## 85 Adelie Dream 37.3 17.8 191 3350 female 2008 ## 86 Adelie Dream 41.3 20.3 194 3550 male 2008 ## 87 Adelie Dream 36.3 19.5 190 3800 male 2008 ## 88 Adelie Dream 36.9 18.6 189 3500 female 2008 ## 89 Adelie Dream 38.3 19.2 189 3950 male 2008 ## 90 Adelie Dream 38.9 18.8 190 3600 female 2008 ## 91 Adelie Dream 35.7 18.0 202 3550 female 2008 ## 92 Adelie Dream 41.1 18.1 205 4300 male 2008 ## 93 Adelie Dream 34.0 17.1 185 3400 female 2008 ## 94 Adelie Dream 39.6 18.1 186 4450 male 2008 ## 95 Adelie Dream 36.2 17.3 187 3300 female 2008 ## 96 Adelie Dream 40.8 18.9 208 4300 male 2008 ## 97 Adelie Dream 38.1 18.6 190 3700 female 2008 ## 98 Adelie Dream 40.3 18.5 196 4350 male 2008 ## 99 Adelie Dream 33.1 16.1 178 2900 female 2008 ## 100 Adelie Dream 43.2 18.5 192 4100 male 2008 ## 101 Adelie Biscoe 35.0 17.9 192 3725 female 2009 ## 102 Adelie Biscoe 41.0 20.0 203 4725 male 2009 ## 103 Adelie Biscoe 37.7 16.0 183 3075 female 2009 ## 104 Adelie Biscoe 37.8 20.0 190 4250 male 2009 ## 105 Adelie Biscoe 37.9 18.6 193 2925 female 2009 ## 106 Adelie Biscoe 39.7 18.9 184 3550 male 2009 ## 107 Adelie Biscoe 38.6 17.2 199 3750 female 2009 ## 108 Adelie Biscoe 38.2 20.0 190 3900 male 2009 ## 109 Adelie Biscoe 38.1 17.0 181 3175 female 2009 ## 110 Adelie Biscoe 43.2 19.0 197 4775 male 2009 ## 111 Adelie Biscoe 38.1 16.5 198 3825 female 2009 ## 112 Adelie Biscoe 45.6 20.3 191 4600 male 2009 ## 113 Adelie Biscoe 39.7 17.7 193 3200 female 2009 ## 114 Adelie Biscoe 42.2 19.5 197 4275 male 2009 ## 115 Adelie Biscoe 39.6 20.7 191 3900 female 2009 ## 116 Adelie Biscoe 42.7 18.3 196 4075 male 2009 ## 117 Adelie Torgersen 38.6 17.0 188 2900 female 2009 ## 118 Adelie Torgersen 37.3 20.5 199 3775 male 2009 ## 119 Adelie Torgersen 35.7 17.0 189 3350 female 2009 ## 120 Adelie Torgersen 41.1 18.6 189 3325 male 2009 ## 121 Adelie Torgersen 36.2 17.2 187 3150 female 2009 ## 122 Adelie Torgersen 37.7 19.8 198 3500 male 2009 ## 123 Adelie Torgersen 40.2 17.0 176 3450 female 2009 ## 124 Adelie Torgersen 41.4 18.5 202 3875 male 2009 ## 125 Adelie Torgersen 35.2 15.9 186 3050 female 2009 ## 126 Adelie Torgersen 40.6 19.0 199 4000 male 2009 ## 127 Adelie Torgersen 38.8 17.6 191 3275 female 2009 ## 128 Adelie Torgersen 41.5 18.3 195 4300 male 2009 ## 129 Adelie Torgersen 39.0 17.1 191 3050 female 2009 ## 130 Adelie Torgersen 44.1 18.0 210 4000 male 2009 ## 131 Adelie Torgersen 38.5 17.9 190 3325 female 2009 ## 132 Adelie Torgersen 43.1 19.2 197 3500 male 2009 ## 133 Adelie Dream 36.8 18.5 193 3500 female 2009 ## 134 Adelie Dream 37.5 18.5 199 4475 male 2009 ## 135 Adelie Dream 38.1 17.6 187 3425 female 2009 ## 136 Adelie Dream 41.1 17.5 190 3900 male 2009 ## 137 Adelie Dream 35.6 17.5 191 3175 female 2009 ## 138 Adelie Dream 40.2 20.1 200 3975 male 2009 ## 139 Adelie Dream 37.0 16.5 185 3400 female 2009 ## 140 Adelie Dream 39.7 17.9 193 4250 male 2009 ## 141 Adelie Dream 40.2 17.1 193 3400 female 2009 ## 142 Adelie Dream 40.6 17.2 187 3475 male 2009 ## 143 Adelie Dream 32.1 15.5 188 3050 female 2009 ## 144 Adelie Dream 40.7 17.0 190 3725 male 2009 ## 145 Adelie Dream 37.3 16.8 192 3000 female 2009 ## 146 Adelie Dream 39.0 18.7 185 3650 male 2009 ## 147 Adelie Dream 39.2 18.6 190 4250 male 2009 ## 148 Adelie Dream 36.6 18.4 184 3475 female 2009 ## 149 Adelie Dream 36.0 17.8 195 3450 female 2009 ## 150 Adelie Dream 37.8 18.1 193 3750 male 2009 ## 151 Adelie Dream 36.0 17.1 187 3700 female 2009 ## 152 Adelie Dream 41.5 18.5 201 4000 male 2009 ## 153 Gentoo Biscoe 46.1 13.2 211 4500 female 2007 ## 154 Gentoo Biscoe 50.0 16.3 230 5700 male 2007 ## 155 Gentoo Biscoe 48.7 14.1 210 4450 female 2007 ## 156 Gentoo Biscoe 50.0 15.2 218 5700 male 2007 ## 157 Gentoo Biscoe 47.6 14.5 215 5400 male 2007 ## 158 Gentoo Biscoe 46.5 13.5 210 4550 female 2007 ## 159 Gentoo Biscoe 45.4 14.6 211 4800 female 2007 ## 160 Gentoo Biscoe 46.7 15.3 219 5200 male 2007 ## 161 Gentoo Biscoe 43.3 13.4 209 4400 female 2007 ## 162 Gentoo Biscoe 46.8 15.4 215 5150 male 2007 ## 163 Gentoo Biscoe 40.9 13.7 214 4650 female 2007 ## 164 Gentoo Biscoe 49.0 16.1 216 5550 male 2007 ## 165 Gentoo Biscoe 45.5 13.7 214 4650 female 2007 ## 166 Gentoo Biscoe 48.4 14.6 213 5850 male 2007 ## 167 Gentoo Biscoe 45.8 14.6 210 4200 female 2007 ## 168 Gentoo Biscoe 49.3 15.7 217 5850 male 2007 ## 169 Gentoo Biscoe 42.0 13.5 210 4150 female 2007 ## 170 Gentoo Biscoe 49.2 15.2 221 6300 male 2007 ## 171 Gentoo Biscoe 46.2 14.5 209 4800 female 2007 ## 172 Gentoo Biscoe 48.7 15.1 222 5350 male 2007 ## 173 Gentoo Biscoe 50.2 14.3 218 5700 male 2007 ## 174 Gentoo Biscoe 45.1 14.5 215 5000 female 2007 ## 175 Gentoo Biscoe 46.5 14.5 213 4400 female 2007 ## 176 Gentoo Biscoe 46.3 15.8 215 5050 male 2007 ## 177 Gentoo Biscoe 42.9 13.1 215 5000 female 2007 ## 178 Gentoo Biscoe 46.1 15.1 215 5100 male 2007 ## 179 Gentoo Biscoe 44.5 14.3 216 4100 <NA> 2007 ## 180 Gentoo Biscoe 47.8 15.0 215 5650 male 2007 ## 181 Gentoo Biscoe 48.2 14.3 210 4600 female 2007 ## 182 Gentoo Biscoe 50.0 15.3 220 5550 male 2007 ## 183 Gentoo Biscoe 47.3 15.3 222 5250 male 2007 ## 184 Gentoo Biscoe 42.8 14.2 209 4700 female 2007 ## 185 Gentoo Biscoe 45.1 14.5 207 5050 female 2007 ## 186 Gentoo Biscoe 59.6 17.0 230 6050 male 2007 ## 187 Gentoo Biscoe 49.1 14.8 220 5150 female 2008 ## 188 Gentoo Biscoe 48.4 16.3 220 5400 male 2008 ## 189 Gentoo Biscoe 42.6 13.7 213 4950 female 2008 ## 190 Gentoo Biscoe 44.4 17.3 219 5250 male 2008 ## 191 Gentoo Biscoe 44.0 13.6 208 4350 female 2008 ## 192 Gentoo Biscoe 48.7 15.7 208 5350 male 2008 ## 193 Gentoo Biscoe 42.7 13.7 208 3950 female 2008 ## 194 Gentoo Biscoe 49.6 16.0 225 5700 male 2008 ## 195 Gentoo Biscoe 45.3 13.7 210 4300 female 2008 ## 196 Gentoo Biscoe 49.6 15.0 216 4750 male 2008 ## 197 Gentoo Biscoe 50.5 15.9 222 5550 male 2008 ## 198 Gentoo Biscoe 43.6 13.9 217 4900 female 2008 ## 199 Gentoo Biscoe 45.5 13.9 210 4200 female 2008 ## 200 Gentoo Biscoe 50.5 15.9 225 5400 male 2008 ## 201 Gentoo Biscoe 44.9 13.3 213 5100 female 2008 ## 202 Gentoo Biscoe 45.2 15.8 215 5300 male 2008 ## 203 Gentoo Biscoe 46.6 14.2 210 4850 female 2008 ## 204 Gentoo Biscoe 48.5 14.1 220 5300 male 2008 ## 205 Gentoo Biscoe 45.1 14.4 210 4400 female 2008 ## 206 Gentoo Biscoe 50.1 15.0 225 5000 male 2008 ## 207 Gentoo Biscoe 46.5 14.4 217 4900 female 2008 ## 208 Gentoo Biscoe 45.0 15.4 220 5050 male 2008 ## 209 Gentoo Biscoe 43.8 13.9 208 4300 female 2008 ## 210 Gentoo Biscoe 45.5 15.0 220 5000 male 2008 ## 211 Gentoo Biscoe 43.2 14.5 208 4450 female 2008 ## 212 Gentoo Biscoe 50.4 15.3 224 5550 male 2008 ## 213 Gentoo Biscoe 45.3 13.8 208 4200 female 2008 ## 214 Gentoo Biscoe 46.2 14.9 221 5300 male 2008 ## 215 Gentoo Biscoe 45.7 13.9 214 4400 female 2008 ## 216 Gentoo Biscoe 54.3 15.7 231 5650 male 2008 ## 217 Gentoo Biscoe 45.8 14.2 219 4700 female 2008 ## 218 Gentoo Biscoe 49.8 16.8 230 5700 male 2008 ## 219 Gentoo Biscoe 46.2 14.4 214 4650 <NA> 2008 ## 220 Gentoo Biscoe 49.5 16.2 229 5800 male 2008 ## 221 Gentoo Biscoe 43.5 14.2 220 4700 female 2008 ## 222 Gentoo Biscoe 50.7 15.0 223 5550 male 2008 ## 223 Gentoo Biscoe 47.7 15.0 216 4750 female 2008 ## 224 Gentoo Biscoe 46.4 15.6 221 5000 male 2008 ## 225 Gentoo Biscoe 48.2 15.6 221 5100 male 2008 ## 226 Gentoo Biscoe 46.5 14.8 217 5200 female 2008 ## 227 Gentoo Biscoe 46.4 15.0 216 4700 female 2008 ## 228 Gentoo Biscoe 48.6 16.0 230 5800 male 2008 ## 229 Gentoo Biscoe 47.5 14.2 209 4600 female 2008 ## 230 Gentoo Biscoe 51.1 16.3 220 6000 male 2008 ## 231 Gentoo Biscoe 45.2 13.8 215 4750 female 2008 ## 232 Gentoo Biscoe 45.2 16.4 223 5950 male 2008 ## 233 Gentoo Biscoe 49.1 14.5 212 4625 female 2009 ## 234 Gentoo Biscoe 52.5 15.6 221 5450 male 2009 ## 235 Gentoo Biscoe 47.4 14.6 212 4725 female 2009 ## 236 Gentoo Biscoe 50.0 15.9 224 5350 male 2009 ## 237 Gentoo Biscoe 44.9 13.8 212 4750 female 2009 ## 238 Gentoo Biscoe 50.8 17.3 228 5600 male 2009 ## 239 Gentoo Biscoe 43.4 14.4 218 4600 female 2009 ## 240 Gentoo Biscoe 51.3 14.2 218 5300 male 2009 ## 241 Gentoo Biscoe 47.5 14.0 212 4875 female 2009 ## 242 Gentoo Biscoe 52.1 17.0 230 5550 male 2009 ## 243 Gentoo Biscoe 47.5 15.0 218 4950 female 2009 ## 244 Gentoo Biscoe 52.2 17.1 228 5400 male 2009 ## 245 Gentoo Biscoe 45.5 14.5 212 4750 female 2009 ## 246 Gentoo Biscoe 49.5 16.1 224 5650 male 2009 ## 247 Gentoo Biscoe 44.5 14.7 214 4850 female 2009 ## 248 Gentoo Biscoe 50.8 15.7 226 5200 male 2009 ## 249 Gentoo Biscoe 49.4 15.8 216 4925 male 2009 ## 250 Gentoo Biscoe 46.9 14.6 222 4875 female 2009 ## 251 Gentoo Biscoe 48.4 14.4 203 4625 female 2009 ## 252 Gentoo Biscoe 51.1 16.5 225 5250 male 2009 ## 253 Gentoo Biscoe 48.5 15.0 219 4850 female 2009 ## 254 Gentoo Biscoe 55.9 17.0 228 5600 male 2009 ## 255 Gentoo Biscoe 47.2 15.5 215 4975 female 2009 ## 256 Gentoo Biscoe 49.1 15.0 228 5500 male 2009 ## 257 Gentoo Biscoe 47.3 13.8 216 4725 <NA> 2009 ## 258 Gentoo Biscoe 46.8 16.1 215 5500 male 2009 ## 259 Gentoo Biscoe 41.7 14.7 210 4700 female 2009 ## 260 Gentoo Biscoe 53.4 15.8 219 5500 male 2009 ## 261 Gentoo Biscoe 43.3 14.0 208 4575 female 2009 ## 262 Gentoo Biscoe 48.1 15.1 209 5500 male 2009 ## 263 Gentoo Biscoe 50.5 15.2 216 5000 female 2009 ## 264 Gentoo Biscoe 49.8 15.9 229 5950 male 2009 ## 265 Gentoo Biscoe 43.5 15.2 213 4650 female 2009 ## 266 Gentoo Biscoe 51.5 16.3 230 5500 male 2009 ## 267 Gentoo Biscoe 46.2 14.1 217 4375 female 2009 ## 268 Gentoo Biscoe 55.1 16.0 230 5850 male 2009 ## 269 Gentoo Biscoe 44.5 15.7 217 4875 <NA> 2009 ## 270 Gentoo Biscoe 48.8 16.2 222 6000 male 2009 ## 271 Gentoo Biscoe 47.2 13.7 214 4925 female 2009 ## 272 Gentoo Biscoe NA NA NA NA <NA> 2009 ## 273 Gentoo Biscoe 46.8 14.3 215 4850 female 2009 ## 274 Gentoo Biscoe 50.4 15.7 222 5750 male 2009 ## 275 Gentoo Biscoe 45.2 14.8 212 5200 female 2009 ## 276 Gentoo Biscoe 49.9 16.1 213 5400 male 2009 ## 277 Chinstrap Dream 46.5 17.9 192 3500 female 2007 ## 278 Chinstrap Dream 50.0 19.5 196 3900 male 2007 ## 279 Chinstrap Dream 51.3 19.2 193 3650 male 2007 ## 280 Chinstrap Dream 45.4 18.7 188 3525 female 2007 ## 281 Chinstrap Dream 52.7 19.8 197 3725 male 2007 ## 282 Chinstrap Dream 45.2 17.8 198 3950 female 2007 ## 283 Chinstrap Dream 46.1 18.2 178 3250 female 2007 ## 284 Chinstrap Dream 51.3 18.2 197 3750 male 2007 ## 285 Chinstrap Dream 46.0 18.9 195 4150 female 2007 ## 286 Chinstrap Dream 51.3 19.9 198 3700 male 2007 ## 287 Chinstrap Dream 46.6 17.8 193 3800 female 2007 ## 288 Chinstrap Dream 51.7 20.3 194 3775 male 2007 ## 289 Chinstrap Dream 47.0 17.3 185 3700 female 2007 ## 290 Chinstrap Dream 52.0 18.1 201 4050 male 2007 ## 291 Chinstrap Dream 45.9 17.1 190 3575 female 2007 ## 292 Chinstrap Dream 50.5 19.6 201 4050 male 2007 ## 293 Chinstrap Dream 50.3 20.0 197 3300 male 2007 ## 294 Chinstrap Dream 58.0 17.8 181 3700 female 2007 ## 295 Chinstrap Dream 46.4 18.6 190 3450 female 2007 ## 296 Chinstrap Dream 49.2 18.2 195 4400 male 2007 ## 297 Chinstrap Dream 42.4 17.3 181 3600 female 2007 ## 298 Chinstrap Dream 48.5 17.5 191 3400 male 2007 ## 299 Chinstrap Dream 43.2 16.6 187 2900 female 2007 ## 300 Chinstrap Dream 50.6 19.4 193 3800 male 2007 ## 301 Chinstrap Dream 46.7 17.9 195 3300 female 2007 ## 302 Chinstrap Dream 52.0 19.0 197 4150 male 2007 ## 303 Chinstrap Dream 50.5 18.4 200 3400 female 2008 ## 304 Chinstrap Dream 49.5 19.0 200 3800 male 2008 ## 305 Chinstrap Dream 46.4 17.8 191 3700 female 2008 ## 306 Chinstrap Dream 52.8 20.0 205 4550 male 2008 ## 307 Chinstrap Dream 40.9 16.6 187 3200 female 2008 ## 308 Chinstrap Dream 54.2 20.8 201 4300 male 2008 ## 309 Chinstrap Dream 42.5 16.7 187 3350 female 2008 ## 310 Chinstrap Dream 51.0 18.8 203 4100 male 2008 ## 311 Chinstrap Dream 49.7 18.6 195 3600 male 2008 ## 312 Chinstrap Dream 47.5 16.8 199 3900 female 2008 ## 313 Chinstrap Dream 47.6 18.3 195 3850 female 2008 ## 314 Chinstrap Dream 52.0 20.7 210 4800 male 2008 ## 315 Chinstrap Dream 46.9 16.6 192 2700 female 2008 ## 316 Chinstrap Dream 53.5 19.9 205 4500 male 2008 ## 317 Chinstrap Dream 49.0 19.5 210 3950 male 2008 ## 318 Chinstrap Dream 46.2 17.5 187 3650 female 2008 ## 319 Chinstrap Dream 50.9 19.1 196 3550 male 2008 ## 320 Chinstrap Dream 45.5 17.0 196 3500 female 2008 ## 321 Chinstrap Dream 50.9 17.9 196 3675 female 2009 ## 322 Chinstrap Dream 50.8 18.5 201 4450 male 2009 ## 323 Chinstrap Dream 50.1 17.9 190 3400 female 2009 ## 324 Chinstrap Dream 49.0 19.6 212 4300 male 2009 ## 325 Chinstrap Dream 51.5 18.7 187 3250 male 2009 ## 326 Chinstrap Dream 49.8 17.3 198 3675 female 2009 ## 327 Chinstrap Dream 48.1 16.4 199 3325 female 2009 ## 328 Chinstrap Dream 51.4 19.0 201 3950 male 2009 ## 329 Chinstrap Dream 45.7 17.3 193 3600 female 2009 ## 330 Chinstrap Dream 50.7 19.7 203 4050 male 2009 ## 331 Chinstrap Dream 42.5 17.3 187 3350 female 2009 ## 332 Chinstrap Dream 52.2 18.8 197 3450 male 2009 ## 333 Chinstrap Dream 45.2 16.6 191 3250 female 2009 ## 334 Chinstrap Dream 49.3 19.9 203 4050 male 2009 ## 335 Chinstrap Dream 50.2 18.8 202 3800 male 2009 ## 336 Chinstrap Dream 45.6 19.4 194 3525 female 2009 ## 337 Chinstrap Dream 51.9 19.5 206 3950 male 2009 ## 338 Chinstrap Dream 46.8 16.5 189 3650 female 2009 ## 339 Chinstrap Dream 45.7 17.0 195 3650 female 2009 ## 340 Chinstrap Dream 55.8 19.8 207 4000 male 2009 ## 341 Chinstrap Dream 43.5 18.1 202 3400 female 2009 ## 342 Chinstrap Dream 49.6 18.2 193 3775 male 2009 ## 343 Chinstrap Dream 50.8 19.0 210 4100 male 2009 ## 344 Chinstrap Dream 50.2 18.7 198 3775 female 2009 ``` --- # Summarize with `summarize()` ### `skewness()`, `kurtosis()` - From `moments` package ```r library(moments) summarize(penguins, skew_mass = skewness(body_mass_g, na.rm = TRUE), kurt_mass = kurtosis(body_mass_g, na.rm = TRUE)) ``` ``` ## # A tibble: 1 × 2 ## skew_mass kurt_mass ## <dbl> <dbl> ## 1 0.468 2.27 ``` .medium[ > 1. Normal distribution, skew = 0, kurtosis = 3* > 2. Remember that it's best to evaluate the distribution **both** visually and statistically ] .footnote[\* **_Excess_ kurtosis** would be 0 for a normal distribution, but this functions measures **kurtosis**] --- # Summarize with `summarize()` ### Confidence Intervals - By hand! - 95% Confidence interval ranges from [mean - (1.96 * SE)] to [mean + (1.96 * SE)] - You can also express this interval as: mean +/- (1.96 * SE) - Standard Errors (SE) can be calculated by SD / sqrt(n) ```r summarize(penguins, mean_mass = mean(body_mass_g, na.rm = TRUE), sd_mass = sd(body_mass_g, na.rm = TRUE), n = n(), se_mass = sd_mass / sqrt(n), # Calculate Standard Error ci_mass = 1.96 * se_mass, # CI margin of error ci_low_mass = mean_mass - ci_mass, # The lower range ci_high_mass = mean_mass + ci_mass) # The upper range ``` --- # Summarize with `summarize()` ### Confidence Intervals - By hand! - 95% Confidence interval ranges from [mean - (1.96 * SE)] to [mean + (1.96 * SE)] - You can also express this interval as: mean +/- (1.96 * SE) - Standard Errors (SE) can be calculated by SD / sqrt(n) ``` ## # A tibble: 1 × 7 ## mean_mass sd_mass n se_mass ci_mass ci_low_mass ci_high_mass ## <dbl> <dbl> <int> <dbl> <dbl> <dbl> <dbl> ## 1 4202. 802. 344 43.2 84.7 4117. 4287. ``` --- # Put it All Together ```r penguins_sp <- group_by(penguins, species) summarize(penguins_sp, mean_mass = mean(body_mass_g, na.rm = TRUE), sd_mass = sd(body_mass_g, na.rm = TRUE), q25_mass = quantile(body_mass_g, probs = 0.25, na.rm = TRUE), median_mass = median(body_mass_g, na.rm = TRUE), q75_mass = quantile(body_mass_g, probs = 0.25, na.rm = TRUE), n = n(), n_no_missing = sum(!is.na(body_mass_g)), skew_mass = skewness(body_mass_g, na.rm = TRUE), kurt_mass = kurtosis(body_mass_g, na.rm = TRUE), se_mass = sd_mass / sqrt(n), ci_mass = 1.96 * se_mass, ci_low_mass = mean_mass - ci_mass, ci_high_mass = mean_mass + ci_mass) ``` --- # Put it All Together ``` ## species mean_mass sd_mass q25_mass median_mass q75_mass n n_no_missing skew_mass kurt_mass ## 1 Adelie 3700.662 458.5661 3350.0 3700 3350.0 152 151 0.28249381 2.405611 ## 2 Chinstrap 3733.088 384.3351 3487.5 3700 3487.5 68 68 0.24194125 3.463681 ## 3 Gentoo 5076.016 504.1162 4700.0 5000 4700.0 124 123 0.06878276 2.257871 ## se_mass ci_mass ci_low_mass ci_high_mass ## 1 37.19462 72.90146 3627.761 3773.564 ## 2 46.60747 91.35065 3641.738 3824.439 ## 3 45.27097 88.73111 4987.285 5164.747 ``` --- # Put it All Together .small[(**Advanced!**)] ### `pivot_longer()` transposes data - from `tidyr` package .small[(part of `tidyverse`)] ```r penguins_long <- pivot_longer(penguins, cols = c(bill_length_mm, bill_depth_mm, flipper_length_mm, body_mass_g), names_to = "measurement", values_to = "values") penguins_long ``` ``` ## # A tibble: 1,376 × 6 ## species island sex year measurement values ## <fct> <fct> <fct> <int> <chr> <dbl> ## 1 Adelie Torgersen male 2007 bill_length_mm 39.1 ## 2 Adelie Torgersen male 2007 bill_depth_mm 18.7 ## 3 Adelie Torgersen male 2007 flipper_length_mm 181 ## 4 Adelie Torgersen male 2007 body_mass_g 3750 ## 5 Adelie Torgersen female 2007 bill_length_mm 39.5 ## 6 Adelie Torgersen female 2007 bill_depth_mm 17.4 ## 7 Adelie Torgersen female 2007 flipper_length_mm 186 ## 8 Adelie Torgersen female 2007 body_mass_g 3800 ## 9 Adelie Torgersen female 2007 bill_length_mm 40.3 ## 10 Adelie Torgersen female 2007 bill_depth_mm 18 ## # … with 1,366 more rows ``` -- </code> function is doing?</small>) --- # Put it All Together .small[(**Advanced!**)] ```r penguins_long_sp <- group_by(penguins_long, species, measurement) summarize(penguins_long_sp, mean = mean(values, na.rm = TRUE), sd = sd(values, na.rm = TRUE), q25 = quantile(values, probs = 0.25, na.rm = TRUE), median = median(values, na.rm = TRUE), q75 = quantile(values, probs = 0.25, na.rm = TRUE), n = n(), n_no_missing = sum(!is.na(values)), skew = skewness(values, na.rm = TRUE), kurt = kurtosis(values, na.rm = TRUE)) ``` --- # Put it All Together .small[(**Advanced!**)] ``` ## `summarise()` has grouped output by 'species'. You can override using the `.groups` argument. ``` ``` ## # A tibble: 12 × 11 ## # Groups: species [3] ## species measurement mean sd q25 median q75 n n_no_missing skew kurt ## <fct> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int> <dbl> <dbl> ## 1 Adelie bill_depth_mm 18.3 1.22 17.5 18.4 17.5 152 151 0.318 2.90 ## 2 Adelie bill_length_mm 38.8 2.66 36.8 38.8 36.8 152 151 0.160 2.81 ## 3 Adelie body_mass_g 3701. 459. 3350 3700 3350 152 151 0.282 2.41 ## 4 Adelie flipper_length_mm 190. 6.54 186 190 186 152 151 0.0865 3.28 ## 5 Chinstrap bill_depth_mm 18.4 1.14 17.5 18.4 17.5 68 68 0.00673 2.10 ## 6 Chinstrap bill_length_mm 48.8 3.34 46.3 49.6 46.3 68 68 -0.0886 2.95 ## 7 Chinstrap body_mass_g 3733. 384. 3488. 3700 3488. 68 68 0.242 3.46 ## 8 Chinstrap flipper_length_mm 196. 7.13 191 196 191 68 68 -0.00926 2.96 ## 9 Gentoo bill_depth_mm 15.0 0.981 14.2 15 14.2 124 123 0.320 2.39 ## 10 Gentoo bill_length_mm 47.5 3.08 45.3 47.3 45.3 124 123 0.643 4.20 ## 11 Gentoo body_mass_g 5076. 504. 4700 5000 4700 124 123 0.0688 2.26 ## 12 Gentoo flipper_length_mm 217. 6.48 212 216 212 124 123 0.390 2.40 ``` --- class: space-list # All Data vs. Variable by Variable ### Depends on what you need - `ggpairs()` and `skim()` - Lots of data quickly summarized and examined - Less easily customized (but still possible!) - `ggplot()` and `summarize()` - Take a bit longer to write out - Very customizable - Can easily include stats not available in `ggpairs()` and `skim()` --- # Wrapping up: Further reading (all **Free**!) - RStudio > Help > Cheatsheets > Data Transformation with dplyr - [R for Data Science](https://r4ds.had.co.nz) - [Data transformation](https://r4ds.had.co.nz/transform.html) - [Exploratory Data Analysis](https://r4ds.had.co.nz/exploratory-data-analysis.html)