class: title-slide, nobar   .footnote[Artwork by [@allison_horst](https://github.com/allisonhorst/stats-illustrations)] ## NRI 7350 # Advanced Linear Models Transformations, Interactions, and Post-hoc tests --- class: section # Getting started (again) Open RStudio Open your NRI project Open a **new** script for today: File > New File > R Script <br> Make sure to load packages at the top: `library(tidyverse)` `library(palmerpenguins)` `library(car)` --- class: section # Side Note ## Messages vs. Warnings vs. Errors --- class: space-list # Messages and Warnings and Errors (Oh my!) - Not all coloured text is a problem - Messages are just helpful information ```r ggplot(data = drop_na(penguins), aes(x = body_mass_g, y = flipper_length_mm)) + stat_smooth() ``` ``` ## `geom_smooth()` using method = 'loess' and formula 'y ~ x' ``` --- class: space-list # Messages and Warnings and Errors (Oh my!) - Not all coloured text is a problem - Messages are just helpful information - Warnings should be considered **FYIs**. They might be a problem, but not always - Your code will run, even with a warning - Warnings always start with **Warning:** ```r ggplot(data = penguins, aes(x = sex, y = body_mass_g)) + geom_boxplot() ``` ``` ## Warning: Removed 2 rows containing non-finite values (stat_boxplot). ``` --- class: space-list # Messages and Warnings and Errors (Oh my!) - Not all coloured text is a problem - Messages are just helpful information - Warnings should be considered **FYIs**. They might be a problem, but not always - Your code will run, even with a warning - Warnings always start with **Warning:** - Errors are always problems 😞 - Your code will stop with an error - Errors always start with **Error:** ```r ggplot(data = Penguins, aes(x = sex, y = body_mass_g)) + geom_boxplot() ``` ``` ## Error in ggplot(data = Penguins, aes(x = sex, y = body_mass_g)): object 'Penguins' not found ``` --  --- class: section # Data Transformations --- class: split-50 # Transformations ## Non-normal residuals .columnl[ ```r m <- lm(sleep_cycle ~ bodywt, data = msleep) d <- data.frame(residuals = residuals(m), std_residuals = rstudent(m), fitted = fitted(m), cooks = cooks.distance(m)) d <- mutate(d, obs = 1:n()) ``` ] .columnr[ ```r ggplot(data = d, aes(sample = std_residuals)) + stat_qq() + stat_qq_line() ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-5-1.png" width="90%" style="display: block; margin: auto;" /> ]  --- # Transformations #### Order of Operations 1. See the need (i.e. non-normal residuals, heteroscedacity, etc.) 2. Figure out which transformation 3. Apply the transformation 4. Check model assumptions 5. Try again as needed --- class: split-55 # Transformations: Common options .columnl[ ### Table of transformations in R ![:spacer 2px]() .small[ .spread[ ```r data_trans <- mutate(data, y_trans = 1/y^2) data_trans <- mutate(data, y_trans = 1/y) data_trans <- mutate(data, y_trans = 1/sqrt(y)) data_trans <- mutate(data, y_trans = log(y)) data_trans <- mutate(data, y_trans = log10(y)) data_trans <- mutate(data, y_trans = sqrt(y)) data_trans <- mutate(data, y_trans = y^2) data_trans <- mutate(data, y_trans = (y^lambda - 1)/lambda) data_trans <- mutate(data, y_trans = asin(sqrt(y/100))) ``` ]]] .columnr[ <table class="table" style="font-size: 20px; margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> Transformation </th> <th style="text-align:left;"> R Code </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;"> Inverse square </td> <td style="text-align:left;font-family: monospace;"> 1/y^2 </td> </tr> <tr> <td style="text-align:left;"> Reciprocal </td> <td style="text-align:left;font-family: monospace;"> 1/y </td> </tr> <tr> <td style="text-align:left;"> Inverse square root </td> <td style="text-align:left;font-family: monospace;"> 1/sqrt(y) </td> </tr> <tr> <td style="text-align:left;"> Naural log (ln) </td> <td style="text-align:left;font-family: monospace;"> log(y) </td> </tr> <tr> <td style="text-align:left;"> Log base 10 </td> <td style="text-align:left;font-family: monospace;"> log10(y) </td> </tr> <tr> <td style="text-align:left;"> Square root </td> <td style="text-align:left;font-family: monospace;"> sqrt(y) </td> </tr> <tr> <td style="text-align:left;"> Square </td> <td style="text-align:left;font-family: monospace;"> y^2 </td> </tr> <tr> <td style="text-align:left;"> Box Cox </td> <td style="text-align:left;font-family: monospace;"> (y^lambda - 1) / lambda </td> </tr> <tr> <td style="text-align:left;"> Arcsine-sqare-root </td> <td style="text-align:left;font-family: monospace;"> asin(sqrt(y/100)) </td> </tr> </tbody> </table> ]  --- class: split-50 # Transformations: How to choose? - Based on what you know (often discipline specific standards for certain data types) - Based on what you see (does it look exponential or logarithmic?) - Based on trial and error (try different transformations and see how it goes) - Based on Box-Cox lambda `\((\lambda)\)` 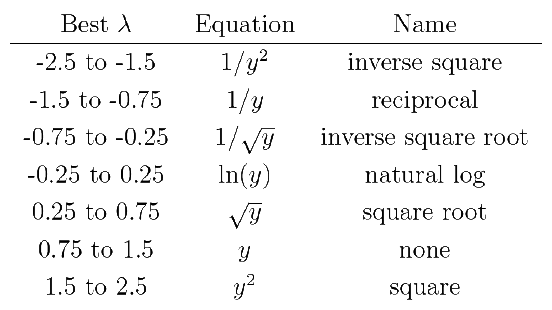 ![:spacer 50px]() .columnl[ .center[ **Can EITHER** **Apply `\(\lambda\)` through Box-Cox transformation** **OR** **Use it to indicate best transformation →** ]] --- class: split-50 # Transformations: Box-Cox ### Finding `\(\lambda\)` ![:spacer 15px]() .columnl[ - **Use a plot of `\(\lambda\)` ** - `boxcox()` function from `MASS` package - Use `boxcox()` directly, otherwise `MASS` interferes with `select()` ```r b <- MASS::boxcox(m) ``` ] .columnr[ <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-8-1.png" width="90%" style="display: block; margin: auto;" /> ] -- .columnl[ - **Get the exact `\(\lambda\)`** ```r b$x[b$y == max(b$y)] ``` ``` ## [1] -0.2626263 ``` ] --- # Apply the transformation ```r msleep_trans <- mutate(msleep, sleep_cycle = (sleep_cycle^(-0.26) - 1) / -0.26) m_trans <- lm(sleep_cycle ~ bodywt, data = msleep_trans) ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-11-1.png" width="100%" style="display: block; margin: auto;" /> --- class: section # Interactions --- # Interactions ### Interaction between Flipper Length and Species Does the effect of Flipper Length on Body Mass depend on Species? (i.e. Are the slopes different?) <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-12-1.png" width="90%" style="display: block; margin: auto;" /> --- # Interactions ### Interaction between Flipper Length and Bill Length Does the effect of Flipper Length on Body Mass depend on Bill Length? (i.e. Does the slope of Flipper Length change with Bill Length?) <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-13-1.png" width="90%" style="display: block; margin: auto;" /> --- # Interactions ### <strong><span style="color:#277F8E">Main Effects</span></strong> only .small[\+] <code class ='r hljs remark-code'>m <- lm(body_mass_g ~ <strong><span style="color:#277F8E">flipper_length_mm </span></strong>+ <strong><span style="color:#277F8E">bill_length_mm</span></strong>, data = penguins)</code> -- ### <strong><span style="color:#277F8E">Main Effects</span></strong> and <strong><span style="color:darkgreen">Interaction</span></strong> .small[\+ :] <code class ='r hljs remark-code'>m <- lm(body_mass_g ~ <strong><span style="color:#277F8E">flipper_length_mm </span></strong>+ <strong><span style="color:#277F8E">bill_length_mm </span></strong>+ <strong><span style="color:darkgreen">flipper_length_mm:bill_length_mm</span></strong>, <br> data = penguins)</code> -- ### *Both* <strong><span style="color:#277F8E">Main Effects</span></strong> and <strong><span style="color:darkgreen">Interaction</span></strong> .small[\* (shortcut)] <code class ='r hljs remark-code'>m <- lm(body_mass_g ~ <strong><span style="color:darkgreen">flipper</span><span style="color:#277F8E">_length</span><span style="color:darkgreen">_mm</span></strong> <strong><span style="color:darkgreen">*</span></strong> <strong><span style="color:darkgreen">bill</span><span style="color:#277F8E">_length</span><span style="color:darkgreen">_mm</span></strong>, data = penguins)</code> -- ![:spacer 20px]() > Don't forget your diagnostic plots! -- </code>) --- # Interpreting Interactions ### Including Correlation Tables <code class ='r hljs remark-code'>summary(m, <span style="background-color:#ffff7f">correlation = TRUE</span>)</code> .small[ ``` ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 5090.5088 2925.3007 1.740 0.082740 . ## flipper_length_mm -7.3085 15.0321 -0.486 0.627145 ## bill_length_mm -229.2424 63.4334 -3.614 0.000347 *** ## flipper_length_mm:bill_length_mm 1.1998 0.3224 3.721 0.000232 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 386.8 on 338 degrees of freedom ## (2 observations deleted due to missingness) ## Multiple R-squared: 0.7694, Adjusted R-squared: 0.7674 ## F-statistic: 375.9 on 3 and 338 DF, p-value: < 2.2e-16 ## *## Correlation of Coefficients: *## (Intercept) flipper_length_mm bill_length_mm *## flipper_length_mm -1.00 *## bill_length_mm -0.99 0.98 *## flipper_length_mm:bill_length_mm 0.99 -0.99 -1.00 ``` ] --  --- layout: true # Interpreting Interactions --- ``` ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 5090.5088 2925.3007 1.740 0.082740 . *## flipper_length_mm -7.3085 15.0321 -0.486 0.627145 ## bill_length_mm -229.2424 63.4334 -3.614 0.000347 *** ## flipper_length_mm:bill_length_mm 1.1998 0.3224 3.721 0.000232 *** ```  --  --  --- ``` ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 5090.5088 2925.3007 1.740 0.082740 . ## flipper_length_mm -7.3085 15.0321 -0.486 0.627145 *## bill_length_mm -229.2424 63.4334 -3.614 0.000347 *** ## flipper_length_mm:bill_length_mm 1.1998 0.3224 3.721 0.000232 *** ```  --  --  --- ``` ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 5090.5088 2925.3007 1.740 0.082740 . ## flipper_length_mm -7.3085 15.0321 -0.486 0.627145 ## bill_length_mm -229.2424 63.4334 -3.614 0.000347 *** *## flipper_length_mm:bill_length_mm 1.1998 0.3224 3.721 0.000232 *** ```  -- ) --  ---  .footnote[Artwork by [@allison_horst](https://github.com/allisonhorst/stats-illustrations)] --- layout: false # Plotting Interactions ### Create new data frame with extremes ```r penguins_new <- expand(penguins, flipper_length_mm = c(min(flipper_length_mm, na.rm = TRUE), max(flipper_length_mm, na.rm = TRUE)), bill_length_mm = c(min(bill_length_mm, na.rm = TRUE), max(bill_length_mm, na.rm = TRUE))) ``` --- # Side Note: `tidyverse` functions ### Create new data frame with extremes <code class ='r hljs remark-code'>penguins_new <- expand(<strong><span style="color:#440154">penguins,</span></strong><br> <strong><span style="color:#277F8E">flipper_length_mm =</span></strong> c(min(<strong><span style="color:deeppink">flipper_length_mm,</span></strong> na.rm = TRUE),<br> max(<strong><span style="color:deeppink">flipper_length_mm,</span></strong> na.rm = TRUE)),<br> <strong><span style="color:#277F8E">bill_length_mm =</span></strong> c(min(<strong><span style="color:deeppink">bill_length_mm,</span></strong> na.rm = TRUE),<br> max(<strong><span style="color:deeppink">bill_length_mm,</span></strong> na.rm = TRUE)))</code> ### `expand()` - from `tidyr` package .small[(part of the `tidyverse`)] - `tidyverse` functions always start with the <span style="color:#440154">**data**</span>, followed by **<span style="color:#277F8E">other arguments</span>** - you can reference any <strong><span style="color:deeppink">column </span></strong>from '<span style="color:#440154">**data**</span>' - `expand()` creates a data frame with all possible combinations of <strong><span style="color:#277F8E">new columns</span></strong> --- # Plotting Interactions ### Create new data frame with extremes ```r penguins_new <- expand(penguins, flipper_length_mm = c(min(flipper_length_mm, na.rm = TRUE), max(flipper_length_mm, na.rm = TRUE)), bill_length_mm = c(min(bill_length_mm, na.rm = TRUE), max(bill_length_mm, na.rm = TRUE))) ``` ### Add predicted y values - Use `predict()` function - `predict()` can be used on most statistical models - Returns predicted `body_mass_g` values for new data ```r penguins_new <- mutate(penguins_new, body_mass_g = predict(m, newdata = penguins_new)) ``` --- # Plotting Interactions ### Small data frame - With values predicted from model - Can plot this to illustrate interactions ```r penguins_new ``` ``` ## # A tibble: 4 × 3 ## flipper_length_mm bill_length_mm body_mass_g ## <int> <dbl> <dbl> ## 1 172 32.1 3099. ## 2 172 59.6 2470. ## 3 231 32.1 4941. ## 4 231 59.6 6258. ``` --- class: split-40 # Plotting Interactions .columnl[ ### Raw data (no model) .small[ ```r ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g, colour = bill_length_mm)) + geom_point(size = 2) ``` ]] .columnr[ .small[ <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-32-1.png" width="100%" style="display: block; margin: auto;" /> ]] --- class: split-40 # Plotting Interactions .columnl[ ### Raw data + Model interaction .small[ ```r ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g, colour = bill_length_mm)) + geom_point(size = 2) + * geom_line(data = penguins_new, * aes(group = bill_length_mm), * size = 2) ``` ]] .columnr[ <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-33-1.png" width="100%" style="display: block; margin: auto;" /> ] --- class: split-50 # Plotting Interactions .columnl[ ### From a Flipper Length perspective .small[ ```r ggplot(data = penguins, aes(x = flipper_length_mm, y = body_mass_g, colour = bill_length_mm, group = bill_length_mm)) + geom_point() + geom_line(data = penguins_new, size = 2) ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-34-1.png" width="90%" style="display: block; margin: auto;" /> ] ] .columnr[ ### From a Bill Length perspective .compact[.small[ ```r ggplot(data = penguins, aes(x = bill_length_mm, y = body_mass_g, colour = flipper_length_mm, group = flipper_length_mm))+ geom_point() + geom_line(data = penguins_new, size = 2) ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-35-1.png" width="90%" style="display: block; margin: auto;" /> ]]] --- # Visualizing Interactions Not what you would present in a paper, but good to think about <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-36-1.png" width="100%" style="display: block; margin: auto;" /> --- class: section # ANOVAs and Post-Hoc Tests --- # Post-Hoc Tests ### From last week... <code class ='r hljs remark-code'>m <- lm(<strong><span style="color:#440154">body_mass_g</span></strong> ~ <strong><span style="color:deeppink">species</span></strong> + <strong><span style="color:deeppink">sex</span></strong>, data = penguins)</code> ```r Anova(m, type = 3) ``` ``` ## Anova Table (Type III tests) ## ## Response: body_mass_g ## Sum Sq Df F value Pr(>F) ## (Intercept) 1154266972 1 11514.96 < 2.2e-16 *** ## species 143401584 2 715.29 < 2.2e-16 *** ## sex 37090262 1 370.01 < 2.2e-16 *** ## Residuals 32979185 329 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` Differences within groups, but how are they different? --- class: split-50 # Post-Hoc Tests .columnl[ - Males significantly bigger than females - Only two groups, so we can use the figure ] .columnr[ - Are Adelie and Chinstrap the same size? - Looks like it, but no statistical support ] <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-39-1.png" width="100%" style="display: block; margin: auto;" /> --  --- # Estimated Marginal Means ### `emmeans()` function from `emmeans` package - Calculates **estimated marginal means** (least-squares means) - Mean response of each factor, adjusting for other factors - Can think of this as a predicted mean response if sample sizes were equal (and controlling for other parameters) ```r library(emmeans) emm_sp <- emmeans(m, specs = "species") emm_sp ``` ``` ## species emmean SE df lower.CL upper.CL ## Adelie 3706 26.2 329 3655 3758 ## Chinstrap 3733 38.4 329 3658 3809 ## Gentoo 5084 29.0 329 5027 5141 ## *## Results are averaged over the levels of: sex ## Confidence level used: 0.95 ``` --- class: split-35, space-list layout: true # Post-Hoc Tests .small[(All pair-wise)] ### `pairs()` function from `emmeans` packages .columnl[ - Compare each combination ```r pairs(emm_sp) ``` ] --- .columnr[ ![:spacer 10px]() .medium[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -26.9 46.5 329 -0.579 0.8313 ## Adelie - Gentoo -1377.9 39.1 329 -35.236 <.0001 ## Chinstrap - Gentoo -1350.9 48.1 329 -28.067 <.0001 ## ## Results are averaged over the levels of: sex ## P value adjustment: tukey method for comparing a family of 3 estimates ``` ]] --- .columnr[ ![:spacer 10px]() .medium[ ``` ## contrast estimate SE df t.ratio p.value *## Adelie - Chinstrap -26.9 46.5 329 -0.579 0.8313 ## Adelie - Gentoo -1377.9 39.1 329 -35.236 <.0001 ## Chinstrap - Gentoo -1350.9 48.1 329 -28.067 <.0001 ## ## Results are averaged over the levels of: sex ## P value adjustment: tukey method for comparing a family of 3 estimates ``` ]] .columnl[ **Differences in Body Mass** .medium[ - Adelie penguins are **not** different from Chinstrap penguins (P value = 0.831) ]] --- .columnr[ ![:spacer 10px]() .medium[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -26.9 46.5 329 -0.579 0.8313 *## Adelie - Gentoo -1377.9 39.1 329 -35.236 <.0001 ## Chinstrap - Gentoo -1350.9 48.1 329 -28.067 <.0001 ## ## Results are averaged over the levels of: sex ## P value adjustment: tukey method for comparing a family of 3 estimates ``` ]] .columnl[ **Differences in Body Mass** .medium[ - Adelie penguins are **not** different from Chinstrap penguins (P value = 0.831) - Adelie penguins **are** on average 1377.9 g lighter than Gentoo penguins (P value = < 0.0001) ]] --- .columnr[ ![:spacer 10px]() .medium[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -26.9 46.5 329 -0.579 0.8313 ## Adelie - Gentoo -1377.9 39.1 329 -35.236 <.0001 *## Chinstrap - Gentoo -1350.9 48.1 329 -28.067 <.0001 ## ## Results are averaged over the levels of: sex ## P value adjustment: tukey method for comparing a family of 3 estimates ``` ]] .columnl[ **Differences in Body Mass** .medium[ - Adelie penguins are **not** different from Chinstrap penguins (P value = 0.831) - Adelie penguins **are** on average 1377.9 g lighter than Gentoo penguins (P value = < 0.0001) - Chinstrap penguins **are** on average 1350.9 g lighter than Gentoo penguins (P value < 0.0001) ]] --  --- layout: false # Post-Hoc Tests .small[(Dunnett's)] ### With `contrast()` function from `emmeans` package - Dunnett's comparison is a type of contrast - Each level compared to control - Use `method = "trt.vs.ctrl"` OR `method = "dunnett"` ```r contrast(emm_sp, method = "dunnett") ``` ``` ## contrast estimate SE df t.ratio p.value ## Chinstrap - Adelie 26.9 46.5 329 0.579 0.7753 ## Gentoo - Adelie 1377.9 39.1 329 35.236 <.0001 ## ## Results are averaged over the levels of: sex ## P value adjustment: dunnettx method for 2 tests ``` -- > Look familiar? --- class: split-50 # Post-Hoc Tests .small[(Dunnett's)] ### Dunnett's treatment vs. control contrasts with `summary()` table .small[ ```r summary(m) ``` ``` ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 3372.39 31.43 107.308 <2e-16 *** *## speciesChinstrap 26.92 46.48 0.579 0.563 *## speciesGentoo 1377.86 39.10 35.236 <2e-16 *** *## sexmale 667.56 34.70 19.236 <2e-16 *** ``` ] ### Dunnett's treatment vs. control contrasts with `emmeans` ![:spacer 5px]() .columnl[ .small[ ```r contrast(emm_sp, method = "dunnett", adjust = "none") ``` ``` ## contrast estimate SE df t.ratio p.value *## Chinstrap - Adelie 26.9 46.5 329 0.579 0.5628 *## Gentoo - Adelie 1377.9 39.1 329 35.236 <.0001 ## ## Results are averaged over the levels of: sex ``` ]] .columnr[ .small[ ```r contrast(emmeans(m, specs = "sex"), method = "dunnett", adjust = "none") ``` ``` ## contrast estimate SE df t.ratio p.value *## male - female 668 34.7 329 19.236 <.0001 ## ## Results are averaged over the levels of: species ``` ]] --- class: split-50 # Post-Hoc Tests and P-Value adjustments .footnote[\* Don't use "Bonferroni, [see this article](https://academic.oup.com/beheco/article/15/6/1044/206216)] .columnl[ ### No adjustment, too liberal? ```r pairs(emm_sp, adjust = "none") ``` .small[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -26.9 46.5 329 -0.579 0.5628 ## Adelie - Gentoo -1377.9 39.1 329 -35.236 <.0001 ## Chinstrap - Gentoo -1350.9 48.1 329 -28.067 <.0001 ## ## Results are averaged over the levels of: sex ``` ] ] .columnr[ ### Extremely conservative\* ```r pairs(emm_sp, adjust = "bonferroni") ``` .small[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -26.9 46.5 329 -0.579 1.0000 ## Adelie - Gentoo -1377.9 39.1 329 -35.236 <.0001 ## Chinstrap - Gentoo -1350.9 48.1 329 -28.067 <.0001 ## ## Results are averaged over the levels of: sex ## P value adjustment: bonferroni method for 3 tests ``` ] ]  --- # Post-Hoc Tests and P-Value adjustments ### Middle of the road: Benjamini Hochberg (FDR, False Discovery Rate)\* ```r pairs(emm_sp, adjust = "fdr") ``` ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -26.9 46.5 329 -0.579 0.5628 ## Adelie - Gentoo -1377.9 39.1 329 -35.236 <.0001 ## Chinstrap - Gentoo -1350.9 48.1 329 -28.067 <.0001 ## ## Results are averaged over the levels of: sex ## P value adjustment: fdr method for 3 tests ``` .footnote[\* [See this article](https://academic.oup.com/beheco/article/15/6/1044/206216)] -- > **No one best method** > What is your question? Are you more concerned about Type 1 or Type 2 error? --- class: medium-table # Post-Hoc Tests ### Test options <table class="table" style="margin-left: auto; margin-right: auto;"> <thead> <tr> <th style="text-align:left;"> Argument </th> <th style="text-align:left;"> P-Value Adjustment </th> </tr> </thead> <tbody> <tr> <td style="text-align:left;font-family: monospace;"> none </td> <td style="text-align:left;"> No P-Value Adjustment (essentially Fisher's LSD) </td> </tr> <tr> <td style="text-align:left;font-family: monospace;"> tukey </td> <td style="text-align:left;"> Tukey's HSD (Honestly significant difference), uses the Studentized range distribution with the number of means in the family. </td> </tr> <tr> <td style="text-align:left;font-family: monospace;"> fdr </td> <td style="text-align:left;"> <a href="http://www.jstor.org/stable/2346101">Benjamini-Hochberg Test or False Discovery Rate Test</a> </td> </tr> <tr> <td style="text-align:left;font-family: monospace;"> bonferroni </td> <td style="text-align:left;"> Bonferroni Correction </td> </tr> <tr> <td style="text-align:left;font-family: monospace;"> scheffe </td> <td style="text-align:left;"> Computes p values from F distribution </td> </tr> <tr> <td style="text-align:left;font-family: monospace;"> mvt </td> <td style="text-align:left;"> Adjusted p values based on the joint normal or t distribution of the linear function </td> </tr> <tr> <td style="text-align:left;font-family: monospace;"> holm </td> <td style="text-align:left;"> <a href="http://www.jstor.org/stable/4615733">Holm Test</a> </td> </tr> <tr> <td style="text-align:left;font-family: monospace;"> hochberg </td> <td style="text-align:left;"> <a href="https://doi.org/10.1093/biomet/75.4.800">Hochberg Test</a> </td> </tr> <tr> <td style="text-align:left;font-family: monospace;"> hommel </td> <td style="text-align:left;"> <a href="https://doi.org/10.1093/biomet/75.2.383">Hommel Test</a> </td> </tr> </tbody> </table> --- # Post-Hoc Tests with Interactions ### Model: 2-way ANOVA ```r m <- lm(body_mass_g ~ species * sex, data = penguins) ``` ### Estimated Marginal Means ```r library(emmeans) m_emms <- emmeans(m, specs = c("species", "sex")) m_emms ``` ``` ## species sex emmean SE df lower.CL upper.CL ## Adelie female 3369 36.2 327 3298 3440 ## Chinstrap female 3527 53.1 327 3423 3632 ## Gentoo female 4680 40.6 327 4600 4760 ## Adelie male 4043 36.2 327 3972 4115 ## Chinstrap male 3939 53.1 327 3835 4043 ## Gentoo male 5485 39.6 327 5407 5563 ## ## Confidence level used: 0.95 ``` --- class: split-35, space-list layout: true # Post-Hoc Tests with Interactions .small[(All pair-wise)] ### `pairs()` function from `emmeans` packages .columnl[ - Compare each combination ```r pairs(m_emms, adjust = "fdr") ``` ] --- .columnr[ ![:spacer 10px]() .small[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie female - Chinstrap female -158 64.2 327 -2.465 0.0152 ## Adelie female - Gentoo female -1311 54.4 327 -24.088 <.0001 ## Adelie female - Adelie male -675 51.2 327 -13.174 <.0001 ## Adelie female - Chinstrap male -570 64.2 327 -8.875 <.0001 ## Adelie female - Gentoo male -2116 53.7 327 -39.425 <.0001 ## Chinstrap female - Gentoo female -1153 66.8 327 -17.246 <.0001 ## Chinstrap female - Adelie male -516 64.2 327 -8.037 <.0001 ## Chinstrap female - Chinstrap male -412 75.0 327 -5.487 <.0001 ## Chinstrap female - Gentoo male -1958 66.2 327 -29.564 <.0001 ## Gentoo female - Adelie male 636 54.4 327 11.691 <.0001 ## Gentoo female - Chinstrap male 741 66.8 327 11.085 <.0001 ## Gentoo female - Gentoo male -805 56.7 327 -14.188 <.0001 ## Adelie male - Chinstrap male 105 64.2 327 1.627 0.1047 ## Adelie male - Gentoo male -1441 53.7 327 -26.855 <.0001 ## Chinstrap male - Gentoo male -1546 66.2 327 -23.345 <.0001 ## ## P value adjustment: fdr method for 15 tests ``` ]] --- .columnr[ ![:spacer 10px]() .small[ ``` ## contrast estimate SE df t.ratio p.value *## Adelie female - Chinstrap female -158 64.2 327 -2.465 0.0152 ## Adelie female - Gentoo female -1311 54.4 327 -24.088 <.0001 ## Adelie female - Adelie male -675 51.2 327 -13.174 <.0001 ## Adelie female - Chinstrap male -570 64.2 327 -8.875 <.0001 ## Adelie female - Gentoo male -2116 53.7 327 -39.425 <.0001 ## Chinstrap female - Gentoo female -1153 66.8 327 -17.246 <.0001 ## Chinstrap female - Adelie male -516 64.2 327 -8.037 <.0001 ## Chinstrap female - Chinstrap male -412 75.0 327 -5.487 <.0001 ## Chinstrap female - Gentoo male -1958 66.2 327 -29.564 <.0001 ## Gentoo female - Adelie male 636 54.4 327 11.691 <.0001 ## Gentoo female - Chinstrap male 741 66.8 327 11.085 <.0001 ## Gentoo female - Gentoo male -805 56.7 327 -14.188 <.0001 ## Adelie male - Chinstrap male 105 64.2 327 1.627 0.1047 ## Adelie male - Gentoo male -1441 53.7 327 -26.855 <.0001 ## Chinstrap male - Gentoo male -1546 66.2 327 -23.345 <.0001 ## ## P value adjustment: fdr method for 15 tests ``` ]] .columnl[ **Differences in Body Mass** .medium[ - Female Adelie penguins **are** on average 158 g smaller than female Chinstrap penguins (P value = 0.015) ]] --- .columnr[ ![:spacer 10px]() .small[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie female - Chinstrap female -158 64.2 327 -2.465 0.0152 ## Adelie female - Gentoo female -1311 54.4 327 -24.088 <.0001 ## Adelie female - Adelie male -675 51.2 327 -13.174 <.0001 ## Adelie female - Chinstrap male -570 64.2 327 -8.875 <.0001 ## Adelie female - Gentoo male -2116 53.7 327 -39.425 <.0001 ## Chinstrap female - Gentoo female -1153 66.8 327 -17.246 <.0001 ## Chinstrap female - Adelie male -516 64.2 327 -8.037 <.0001 ## Chinstrap female - Chinstrap male -412 75.0 327 -5.487 <.0001 ## Chinstrap female - Gentoo male -1958 66.2 327 -29.564 <.0001 ## Gentoo female - Adelie male 636 54.4 327 11.691 <.0001 ## Gentoo female - Chinstrap male 741 66.8 327 11.085 <.0001 ## Gentoo female - Gentoo male -805 56.7 327 -14.188 <.0001 *## Adelie male - Chinstrap male 105 64.2 327 1.627 0.1047 ## Adelie male - Gentoo male -1441 53.7 327 -26.855 <.0001 ## Chinstrap male - Gentoo male -1546 66.2 327 -23.345 <.0001 ## ## P value adjustment: fdr method for 15 tests ``` ]] .columnl[ **Differences in Body Mass** .medium[ - Female Adelie penguins **are** on average 158 g smaller than female Chinstrap penguins (P value = 0.015) - Male Adelie penguins are **not** different from male Chinstrap penguins (P value = 0.105) ]] --- .columnr[ ![:spacer 10px]() .small[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie female - Chinstrap female -158 64.2 327 -2.465 0.0152 ## Adelie female - Gentoo female -1311 54.4 327 -24.088 <.0001 *## Adelie female - Adelie male -675 51.2 327 -13.174 <.0001 ## Adelie female - Chinstrap male -570 64.2 327 -8.875 <.0001 ## Adelie female - Gentoo male -2116 53.7 327 -39.425 <.0001 ## Chinstrap female - Gentoo female -1153 66.8 327 -17.246 <.0001 ## Chinstrap female - Adelie male -516 64.2 327 -8.037 <.0001 ## Chinstrap female - Chinstrap male -412 75.0 327 -5.487 <.0001 ## Chinstrap female - Gentoo male -1958 66.2 327 -29.564 <.0001 ## Gentoo female - Adelie male 636 54.4 327 11.691 <.0001 ## Gentoo female - Chinstrap male 741 66.8 327 11.085 <.0001 ## Gentoo female - Gentoo male -805 56.7 327 -14.188 <.0001 ## Adelie male - Chinstrap male 105 64.2 327 1.627 0.1047 ## Adelie male - Gentoo male -1441 53.7 327 -26.855 <.0001 ## Chinstrap male - Gentoo male -1546 66.2 327 -23.345 <.0001 ## ## P value adjustment: fdr method for 15 tests ``` ]] .columnl[ **Differences in Body Mass** .medium[ - Female Adelie penguins **are** on average 158 g smaller than female Chinstrap penguins (P value = 0.015) - Male Adelie penguins are **not** different from male Chinstrap penguins (P value = 0.105) - Female Adelie penguins **are** on average 675 g lighter than male Adelie penguins (P value = 0) ]] --- layout: false class: split-70, space-list # Post-Hoc Tests with Interactions .columnl[ ### Model: ANCOVA ```r m <- lm(body_mass_g ~ flipper_length_mm * species, data = penguins) ``` ### Estimated Marginal Means - Here use `emtrends()` function from `emmeans` package ```r m_emms <- emtrends(m, specs = "species", var = "flipper_length_mm") m_emms ``` .small[ ``` ## species flipper_length_mm.trend SE df lower.CL upper.CL ## Adelie 32.8 4.63 336 23.7 41.9 ## Chinstrap 34.6 6.35 336 22.1 47.1 ## Gentoo 54.6 5.17 336 44.4 64.8 ## ## Confidence level used: 0.95 ``` ] These are the effects of Flipper Length on Body Mass in each Species. ] .columnr[ .medium[ **On average...** - Adelie: Body Mass increases by 32.8 g for each 1 mm increase in Flipper Length. - Chinstrap: Body Mass increases by 34.6 g for each 1 mm increase in Flipper Length. - Gentoo: Body Mass increases by 54.6 g for each 1 mm increase in Flipper Length. ]] --  --- class: split-35, space-list layout: true # Post-Hoc Tests with Interactions .small[(All pair-wise)] ### `pairs()` function from `emmeans` packages .columnl[ - Compare among species ```r pairs(m_emms, adjust = "fdr") ``` ] --- .columnr[ ![:spacer 10px]() .small[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -1.74 7.86 336 -0.222 0.8247 ## Adelie - Gentoo -21.79 6.94 336 -3.139 0.0055 ## Chinstrap - Gentoo -20.05 8.19 336 -2.448 0.0223 ## ## P value adjustment: fdr method for 3 tests ``` ]] --- .columnr[ ![:spacer 10px]() .small[ ``` ## contrast estimate SE df t.ratio p.value *## Adelie - Chinstrap -1.74 7.86 336 -0.222 0.8247 ## Adelie - Gentoo -21.79 6.94 336 -3.139 0.0055 ## Chinstrap - Gentoo -20.05 8.19 336 -2.448 0.0223 ## ## P value adjustment: fdr method for 3 tests ``` ]] ![:spacer 100px]() **Differences in the effect of Flipper Length on Body Mass** .medium[ - The effect of Flipper Length on Body Mass is **not** different between Adelie and Chinstrap penguins (P value = 0.825) ] --- .columnr[ ![:spacer 10px]() .small[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -1.74 7.86 336 -0.222 0.8247 *## Adelie - Gentoo -21.79 6.94 336 -3.139 0.0055 ## Chinstrap - Gentoo -20.05 8.19 336 -2.448 0.0223 ## ## P value adjustment: fdr method for 3 tests ``` ]] ![:spacer 100px]() **Differences in the effect of Flipper Length on Body Mass** .medium[ - The effect of Flipper Length on Body Mass is **not** different between Adelie and Chinstrap penguins (P value = 0.825) - The effect of Flipper Length on Body Mass **is significantly less** in Adelie than in Gentoo penguins (Body Mass increases 21.8 g less per 1mm increase in Flipper Length; P value = 0.006) ] --- .columnr[ ![:spacer 10px]() .small[ ``` ## contrast estimate SE df t.ratio p.value ## Adelie - Chinstrap -1.74 7.86 336 -0.222 0.8247 ## Adelie - Gentoo -21.79 6.94 336 -3.139 0.0055 *## Chinstrap - Gentoo -20.05 8.19 336 -2.448 0.0223 ## ## P value adjustment: fdr method for 3 tests ``` ]] ![:spacer 100px]() **Differences in the effect of Flipper Length on Body Mass** .medium[ - The effect of Flipper Length on Body Mass is **not** different between Adelie and Chinstrap penguins (P value = 0.825) - The effect of Flipper Length on Body Mass **is significantly less** in Adelie than in Gentoo penguins (Body Mass increases 21.8 g less per 1mm increase in Flipper Length; P value = 0.006) - The effect of Flipper Length on Body Mass **is significantly less** in Chinstrap than in Gentoo penguins (Body Mass increases 20 g less per 1mm increase in Flipper Length; P value = 0.022) ] --  --- layout: false class: split-40, space-list # Visualizing Interactions ```r emmip(m, species ~ flipper_length_mm, cov.reduce = range) ``` .columnl[ .medium[ - Here, **Linear prediction** = **y** = **Body Mass** - Adelie and Chinstrap penguins have similar effects (i.e. slopes are **not** significantly different) - Gentoo has a larger effect (i.e. slope **is** significantly larger) than either Adelie or Chinstrap penguins ]] .columnr[ <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-66-1.png" width="100%" style="display: block; margin: auto;" /> ] --- # Homework (Practice)* Consider bill depth your response variable and species and year your predictor variables 1\. Convert year to a categorical variable and remove Gentoo penguins ```r penguins_sub <- mutate(penguins, year = factor(year)) penguins_sub <- filter(penguins_sub, species != "Gentoo") ``` 2\. Create a figure comparing bill depth to species and year. Think about how to best tease apart the relationships. 3\. Model the relationship between bill depth and the interaction between species and year. 4\. Check diagnostics 5\. How does the relationship of species and bill depth change among years? .footnote[\* Not to be handed in, answers posted in these slides next week] --- exclude: FALSE # Homework (Practice) Answers 2\. Create a figure comparing body mass to bill depth and year ```r ggplot(penguins_sub, aes(x = bill_depth_mm, y = body_mass_g, colour = year)) + geom_point() + stat_smooth(method = "lm", se = FALSE) ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-68-1.png" width="70%" style="display: block; margin: auto;" /> --- exclude: FALSE class: split-50 # Homework (Practice) Answers 3\. Model the relationship between body mass and the interaction between bill depth and year. ```r m <- lm(body_mass_g ~ bill_depth_mm * year, data = penguins_sub) ``` --- exclude: FALSE # Homework (Practice) Answers 4\. Check your model diagnostics ```r d <- data.frame(residuals = residuals(m), std_residuals = rstudent(m), fitted = fitted(m), cooks = cooks.distance(m)) d <- mutate(d, obs = 1:n()) d ``` ``` ## residuals std_residuals fitted cooks obs ## 1 48.767953 0.139759238 3701.232 4.422779e-05 1 ## 2 334.691697 0.969259197 3465.308 4.778239e-03 2 ## 3 -324.196185 -0.933081958 3574.196 2.640006e-03 3 ## 5 -360.119929 -1.036704430 3810.120 3.161623e-03 4 ## 6 -396.043674 -1.160886854 4046.044 1.204535e-02 5 ## 7 87.099776 0.250606023 3537.900 2.251416e-04 6 ## 8 810.436130 2.363323737 3864.564 2.057554e-02 7 ## 9 -117.344165 -0.336917273 3592.344 3.208236e-04 8 ## 10 276.548248 0.803388875 3973.452 4.139380e-03 9 ## 11 -110.864362 -0.321859240 3410.864 6.887368e-04 10 ## 12 252.839677 0.732531214 3447.160 2.988740e-03 11 ## 13 -301.604264 -0.870950455 3501.604 3.229564e-03 12 ## 14 -354.931556 -1.054939693 4154.932 1.568554e-02 13 ## 15 263.216425 0.779297638 4136.784 7.985436e-03 14 ## 16 162.099776 0.466568506 3537.900 7.798057e-04 15 ## 17 -305.675988 -0.878044715 3755.676 1.884339e-03 16 ## 18 435.808346 1.281026324 4064.192 1.586371e-02 17 ## 19 -321.788106 -0.924333210 3646.788 2.032933e-03 18 ## 20 -9.375497 -0.028041477 4209.375 1.370199e-05 19 ## 21 -228.640126 -0.656327379 3628.640 1.074801e-03 20 ## 22 -101.232047 -0.290155114 3701.232 1.905734e-04 21 ## 23 8.028051 0.023038881 3791.972 1.461514e-06 22 ## 24 357.655835 1.029184148 3592.344 2.980395e-03 23 ## 25 370.987658 1.078009849 3429.012 7.047852e-03 24 ## 26 62.471992 0.179085488 3737.528 7.552857e-05 25 ## 27 -133.084067 -0.381513303 3683.084 3.303557e-04 26 ## 28 -356.048205 -1.025953499 3556.048 3.453529e-03 27 ## 29 -533.084067 -1.536151293 3683.084 5.300552e-03 28 ## 30 212.471992 0.609570790 3737.528 8.736630e-04 29 ## 31 -88.272441 -0.258174262 3338.272 6.182483e-04 30 ## 32 307.655835 0.884730440 3592.344 2.205330e-03 31 ## 33 -237.900224 -0.685149423 3537.900 1.679621e-03 32 ## 34 162.471992 0.465953919 3737.528 5.108549e-04 33 ## 35 -67.716382 -0.196897903 3392.716 2.808596e-04 34 ## 36 13.216425 0.039073721 4136.784 2.013264e-05 35 ## 37 12.844208 0.037145335 3937.156 7.444577e-06 36 ## 38 -114.936087 -0.329499854 3664.936 2.509617e-04 37 ## 39 -510.119929 -1.472281952 3810.120 6.343958e-03 38 ## 40 876.176031 2.551501243 3773.824 1.633167e-02 39 ## 41 -424.196185 -1.222684981 3574.196 4.519836e-03 40 ## 42 253.211894 0.726791217 3646.788 1.258784e-03 41 ## 43 -564.936087 -1.629254760 3664.936 6.063081e-03 42 ## 44 517.288149 1.498365968 3882.712 9.171684e-03 43 ## 45 -374.568401 -1.094081198 3374.568 9.380741e-03 44 ## 46 880.619972 2.562562573 3719.380 1.460218e-02 45 ## 47 -330.675988 -0.950150633 3755.676 2.205168e-03 46 ## 48 -762.528008 -2.210789611 3737.528 1.125258e-02 47 ## 49 -106.048205 -0.304889117 3556.048 3.063741e-04 48 ## 50 -4.931556 -0.014619423 4154.932 3.028155e-06 49 ## 51 -106.584816 -0.306245393 3606.585 2.895658e-04 50 ## 52 370.510222 1.067755506 3929.490 3.706662e-03 51 ## 53 -210.402309 -0.604311787 3660.402 9.998535e-04 52 ## 54 -40.942259 -0.118418323 4090.942 7.607157e-05 53 ## 55 -814.219803 -2.365854800 3714.220 1.389170e-02 54 ## 56 -148.763537 -0.427056698 3848.764 4.948293e-04 55 ## 57 -2.767322 -0.007961092 3552.767 2.274088e-07 56 ## 58 -102.581031 -0.294699802 3902.581 2.644484e-04 57 ## 59 -460.588600 -1.347050783 3310.589 1.406919e-02 58 ## 60 -233.307271 -0.672372159 3983.307 1.727471e-03 59 ## 61 -241.314841 -0.699974231 3391.315 2.949859e-03 60 ## 62 -121.482208 -0.365064049 4521.482 2.538871e-03 61 ## 63 181.776412 0.526234083 3418.224 1.527973e-03 62 ## 64 308.871450 0.887396567 3741.129 1.966182e-03 63 ## 65 -595.132335 -1.731540709 3445.132 1.495024e-02 64 ## 66 262.688944 0.754592546 3687.311 1.491376e-03 65 ## 67 147.046387 0.432083005 3202.954 2.020754e-03 66 ## 68 116.692729 0.336030195 3983.307 4.321574e-04 67 ## 69 -260.588600 -0.759917060 3310.589 4.503535e-03 68 ## 70 385.966488 1.118101105 4064.034 6.171361e-03 69 ## 71 -356.398524 -1.027669655 3956.399 3.709107e-03 70 ## 72 105.053957 0.301354913 3794.946 2.301925e-04 71 ## 73 77.958919 0.224982615 3472.041 2.342879e-04 72 ## 74 220.510222 0.634377122 3929.490 1.312925e-03 73 ## 75 147.232678 0.423741436 3552.767 6.437190e-04 74 ## 76 428.145210 1.232551969 3821.855 3.932187e-03 75 ## 77 335.593906 0.976054439 3364.406 6.246589e-03 76 ## 78 -164.033512 -0.474042597 4064.034 1.114672e-03 77 ## 79 373.955134 1.104200180 3176.045 1.419206e-02 78 ## 80 16.692729 0.048056107 3983.307 8.843191e-06 79 ## 81 -272.041081 -0.786136391 3472.041 2.852905e-03 80 ## 82 1120.323931 3.302224176 3579.676 3.442429e-02 81 ## 83 -102.581031 -0.294699802 3902.581 2.644484e-04 82 ## 84 135.966488 0.392866186 4064.034 7.658532e-04 83 ## 85 -283.493562 -0.815185076 3633.494 1.918955e-03 84 ## 86 -756.212233 -2.246267180 4306.212 5.253510e-02 85 ## 87 -290.942259 -0.842880555 4090.942 3.841425e-03 86 ## 88 -348.763537 -1.003140291 3848.764 2.719722e-03 87 ## 89 -60.216018 -0.173536575 4010.216 1.255075e-04 88 ## 90 -302.581031 -0.870643947 3902.581 2.300860e-03 89 ## 91 -137.311056 -0.394051295 3687.311 4.074877e-04 90 ## 92 585.780197 1.691347481 3714.220 7.190208e-03 91 ## 93 -45.132335 -0.130399180 3445.132 8.597985e-05 92 ## 94 735.780197 2.132777907 3714.220 1.134405e-02 93 ## 95 -198.949828 -0.573882881 3498.950 1.395194e-03 94 ## 96 370.510222 1.067755506 3929.490 3.706662e-03 95 ## 97 -148.763537 -0.427056698 3848.764 4.948293e-04 96 ## 98 528.145210 1.523283695 3821.855 5.983547e-03 97 ## 99 -276.044866 -0.814029943 3176.045 7.733314e-03 98 ## 100 278.145210 0.799076315 3821.855 1.659568e-03 99 ## 101 100.714641 0.288745028 3624.285 1.959280e-04 100 ## 102 638.988589 1.876067369 4086.011 2.696129e-02 101 ## 103 -131.533217 -0.385740517 3206.533 1.511606e-03 102 ## 104 163.988589 0.477779114 4086.011 1.775749e-03 103 ## 105 -853.194043 -2.482177763 3778.194 1.514210e-02 104 ## 106 -294.154908 -0.846428232 3844.155 2.207370e-03 105 ## 107 279.623325 0.806068978 3470.377 2.437255e-03 106 ## 108 -186.011411 -0.542026034 4086.011 2.284723e-03 107 ## 109 -251.402766 -0.725997111 3426.403 2.348496e-03 108 ## 110 908.858137 2.655872979 3866.142 2.283374e-02 109 ## 111 508.532009 1.484473475 3316.468 1.492846e-02 110 ## 112 448.027724 1.317630968 4151.972 1.695814e-02 111 ## 113 -380.311450 -1.093966776 3580.311 3.082509e-03 112 ## 114 298.923363 0.865317750 3976.077 3.820859e-03 113 ## 115 -339.920095 -1.007337128 4239.920 1.331031e-02 114 ## 116 362.766821 1.042213785 3712.233 2.441720e-03 115 ## 117 -526.402766 -1.526573747 3426.403 1.029641e-02 116 ## 118 -420.946185 -1.242929531 4195.946 1.752520e-02 117 ## 119 -76.402766 -0.220386235 3426.403 2.169041e-04 118 ## 120 -453.194043 -1.304925242 3778.194 4.272272e-03 119 ## 121 -320.376675 -0.923991402 3470.377 3.199454e-03 120 ## 122 -542.037501 -1.582318771 4042.038 1.635500e-02 121 ## 123 23.597234 0.068059930 3426.403 2.069054e-05 122 ## 124 118.792912 0.340657084 3756.207 2.791582e-04 123 ## 125 -134.546262 -0.395455957 3184.546 1.711493e-03 124 ## 126 133.858137 0.384946503 3866.142 4.953069e-04 125 ## 127 -283.324495 -0.814366140 3558.324 1.823832e-03 126 ## 128 587.766821 1.695703862 3712.233 6.409894e-03 127 ## 129 -398.389720 -1.151384777 3448.390 5.398297e-03 128 ## 130 353.727686 1.016177755 3646.272 2.342713e-03 129 ## 131 -299.285359 -0.859363928 3624.285 1.730144e-03 130 ## 132 -410.115772 -1.184990512 3910.116 5.513373e-03 131 ## 133 -256.207088 -0.735448926 3756.207 1.298529e-03 132 ## 134 718.792912 2.081639551 3756.207 1.022061e-02 133 ## 135 -133.324495 -0.382751853 3558.324 4.038652e-04 134 ## 136 363.662460 1.046965117 3536.338 3.227747e-03 135 ## 137 -361.337540 -1.040237518 3536.338 3.186609e-03 136 ## 138 -132.998366 -0.388132888 4107.998 1.269698e-03 137 ## 139 83.532009 0.242617343 3316.468 4.027952e-04 138 ## 140 625.714641 1.807311495 3624.285 7.562474e-03 139 ## 141 -48.389720 -0.139422137 3448.390 7.964285e-05 140 ## 142 4.623325 0.013307268 3470.377 6.662894e-07 141 ## 143 -46.598443 -0.138285548 3096.598 2.779542e-04 142 ## 144 298.597234 0.862725141 3426.403 3.312998e-03 143 ## 145 -382.428856 -1.108905998 3382.429 6.492051e-03 144 ## 146 -150.180998 -0.431089084 3800.181 4.983110e-04 145 ## 147 471.805957 1.358973883 3778.194 4.630387e-03 146 ## 148 -259.220133 -0.743923074 3734.220 1.278956e-03 147 ## 149 -152.298404 -0.436877366 3602.298 4.680253e-04 148 ## 150 81.740731 0.234255827 3668.259 1.229351e-04 149 ## 151 251.610280 0.725815139 3448.390 2.153264e-03 150 ## 152 243.792912 0.699729317 3756.207 1.175741e-03 151 ## 153 -56.048205 -0.161113414 3556.048 8.557933e-05 152 ## 154 53.584110 0.154107334 3846.416 8.240076e-05 153 ## 155 -141.971949 -0.407589840 3791.972 4.570755e-04 154 ## 156 -176.232047 -0.505326736 3701.232 5.775585e-04 155 ## 157 -175.859831 -0.507591631 3900.860 1.161378e-03 156 ## 158 412.099776 1.189479402 3537.900 5.039951e-03 157 ## 159 -360.492146 -1.036831499 3610.492 2.830561e-03 158 ## 160 139.507854 0.400384111 3610.492 4.239144e-04 159 ## 161 412.471992 1.186241818 3737.528 3.292528e-03 160 ## 162 -219.007811 -0.633120376 3919.008 1.974519e-03 161 ## 163 262.099776 0.755022535 3537.900 2.038707e-03 162 ## 164 -216.599733 -0.629911972 3991.600 2.776792e-03 163 ## 165 252.839677 0.732531214 3447.160 2.988740e-03 164 ## 166 457.655835 1.319044324 3592.344 4.880016e-03 165 ## 167 164.135638 0.476654190 3410.864 1.509646e-03 166 ## 168 185.436130 0.534126290 3864.564 1.077215e-03 167 ## 169 -637.155792 -1.857578067 3937.156 1.831963e-02 168 ## 170 162.099776 0.466568506 3537.900 7.798057e-04 169 ## 171 -233.084067 -0.668658935 3683.084 1.013339e-03 170 ## 172 789.507854 2.292925577 3610.492 1.357669e-02 171 ## 173 152.839677 0.442454392 3447.160 1.092119e-03 172 ## 174 -83.456283 -0.240881196 3483.456 2.710030e-04 173 ## 175 -420.124460 -1.235641689 3320.124 1.522330e-02 174 ## 176 -28.267910 -0.081229579 3828.268 2.108322e-05 175 ## 177 -256.048205 -0.736920552 3556.048 1.786031e-03 176 ## 178 394.324012 1.134053665 3755.676 3.135760e-03 177 ## 179 -394.946043 -1.136131909 3794.946 3.253433e-03 178 ## 180 -156.398524 -0.450068488 3956.399 7.142707e-04 179 ## 181 66.506438 0.190956504 3633.494 1.056102e-04 180 ## 182 324.514007 0.948505777 4225.486 7.488538e-03 181 ## 183 -110.588600 -0.322134112 3310.589 8.110796e-04 182 ## 184 -140.755967 -0.418959464 4440.756 2.717388e-03 183 ## 185 12.502653 0.036343840 3337.497 9.485374e-06 184 ## 186 197.418969 0.567469229 3902.581 9.794551e-04 185 ## 187 -248.763537 -0.714679918 3848.764 1.383681e-03 186 ## 188 535.593906 1.563185572 3364.406 1.591058e-02 187 ## 189 81.962703 0.235073040 3768.037 1.382365e-04 188 ## 190 386.152780 1.149092310 4413.847 1.892325e-02 189 ## 191 -610.588600 -1.791561687 3310.589 2.472522e-02 190 ## 192 301.422754 0.879060378 4198.577 5.916830e-03 191 ## 193 -140.942259 -0.407797205 4090.942 9.014907e-04 192 ## 194 97.232678 0.279772672 3552.767 2.807452e-04 193 ## 195 -433.307271 -1.252029577 3983.307 5.958628e-03 194 ## 196 81.776412 0.236615601 3418.224 3.092408e-04 195 ## 197 50.714641 0.145375602 3624.285 4.967958e-05 196 ## 198 693.792912 2.007836951 3756.207 9.522019e-03 197 ## 199 -224.285359 -0.643518754 3624.285 9.716573e-04 198 ## 200 301.936408 0.875239241 3998.064 4.259698e-03 199 ## 201 -550.180998 -1.587942987 3800.181 6.687776e-03 200 ## 202 182.636370 0.525559848 3492.364 9.548793e-04 201 ## 203 30.518964 0.088780020 3294.481 5.856609e-05 202 ## 204 83.858137 0.241106334 3866.142 1.943906e-04 203 ## 205 107.636370 0.309606008 3492.364 3.316592e-04 204 ## 206 29.949453 0.086785311 4020.051 4.576638e-05 205 ## 207 -142.363630 -0.409565134 3492.364 5.801924e-04 206 ## 208 -372.167953 -1.071302590 3822.168 3.277458e-03 207 ## 209 -88.454946 -0.256515706 3338.455 4.141830e-04 208 ## 210 -14.024456 -0.040767382 4064.024 1.193192e-05 209 ## 211 -22.167953 -0.063640053 3822.168 1.162814e-05 210 ## 212 -429.089682 -1.242911432 3954.090 7.202752e-03 211 ## 213 -26.076637 -0.075354214 3976.077 2.907666e-05 212 ## 214 333.532009 0.970754152 3316.468 6.421751e-03 213 ## 215 223.597234 0.645533050 3426.403 1.857730e-03 214 ## 216 -42.037501 -0.122002095 4042.038 9.837069e-05 215 ## 217 -268.259269 -0.769762045 3668.259 1.324062e-03 216 ## 218 84.753776 0.242888244 3690.246 1.317605e-04 217 ## 219 233.858137 0.673007339 3866.142 1.511784e-03 218 ## 220 -25.180998 -0.072250367 3800.181 1.400930e-05 219 ``` --- exclude: FALSE # Homework (Practice) Answers 4\. Check your model diagnostics - Normality ```r ggplot(data = d, aes(sample = std_residuals)) + stat_qq() + stat_qq_line() ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-71-1.png" width="50%" style="display: block; margin: auto;" />  --- exclude: FALSE # Homework (Practice) Answers 4\. Check your model diagnostics - Heteroscedasticity ```r ggplot(d, aes(x = fitted, y = std_residuals)) + geom_point() + geom_hline(yintercept = 0) ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-72-1.png" width="50%" style="display: block; margin: auto;" />  --- exclude: FALSE # Homework (Practice) Answers 4\. Check your model diagnostics - Influence (Cook's d) ```r ggplot(d, aes(x = obs, y = cooks)) + geom_point() + geom_hline(yintercept = 4/nrow(penguins), linetype = "dashed") ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-73-1.png" width="50%" style="display: block; margin: auto;" />  --- class: split-50 exclude: FALSE # Homework (Practice) Answers 5\. How does the relationship of body mass and bill depth change among years? .columnl[ .small[ ```r em <- emtrends(m, specs = "year", var = "bill_depth_mm") pairs(em, adjust = "fdr") ``` ``` ## contrast estimate SE df t.ratio p.value ## 2007 - 2008 -87.6 50.7 213 -1.727 0.2569 ## 2007 - 2009 -38.4 48.9 213 -0.785 0.4331 ## 2008 - 2009 49.2 49.8 213 0.988 0.4331 ## ## P value adjustment: fdr method for 3 tests ``` ] .medium[ > No significant differences in the slopes among the years. > i.e. the relationship between bill depth and body mass did not change among years. ]] .columnr[ .small[ ```r emmip(m, year ~ bill_depth_mm, cov.reduce = range) ``` <img src="5 Interactions and Post-Hoc tests - answers_files/figure-html/unnamed-chunk-75-1.png" width="90%" style="display: block; margin: auto;" /> ]] ![:spacer 140px]()