class: title-slide, nobar 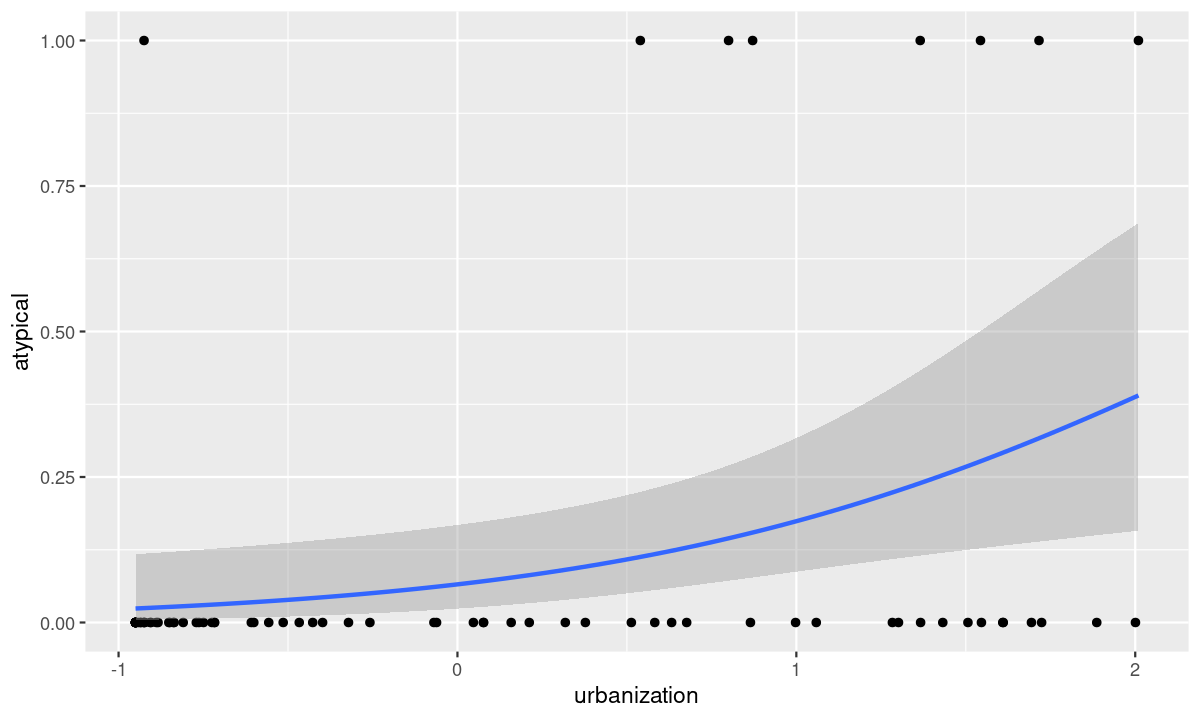  ## NRI 7350 # Even more stats... Generalized linear models, Other advanced models, Non-parametric stats --- class: section # Getting started (again) Open RStudio Open your NRI project Open a **new** script for today: File > New File > R Script <br> Make sure to load packages at the top: `library(tidyverse)` `library(DHARMa)` --- class: section # Reference Material This lecture covers A HUGE subject area It is not comprehensive It is a place to start, with references to guide you later on --- class: section # Diagnostics for complex models ### Introducing DHARMa [DHARMa Tutorial](https://cran.r-project.org/web/packages/DHARMa/vignettes/DHARMa.html) .small[(Many great examples of model checking)] --- class: space-list # DHARMa Package ### *Simulated* Residuals 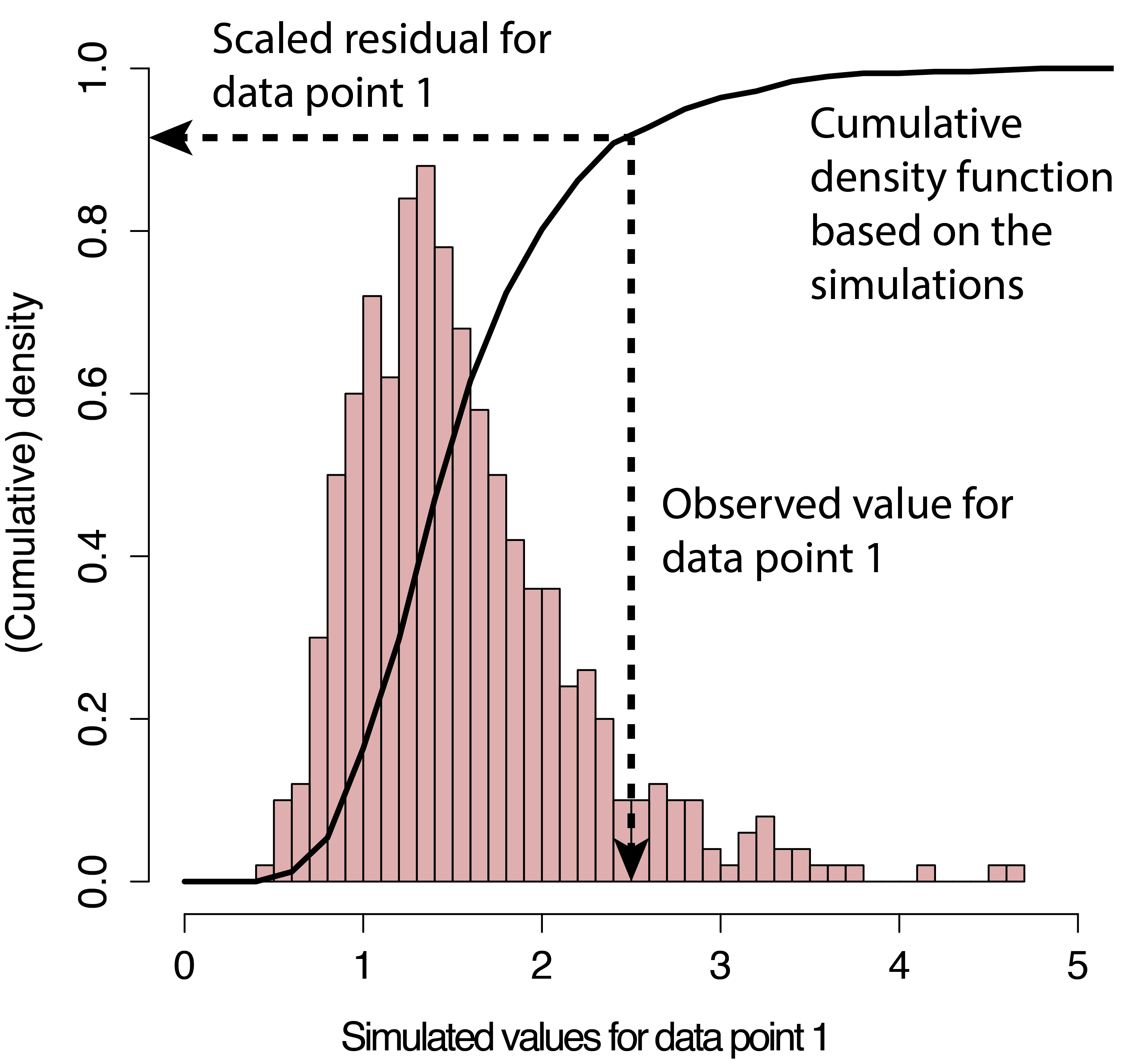 - **If** your data perfectly matched your model,<br>what would the values look like? - Pink histogram values show repeatedly<br>simulated values -- - How do your *actual* values compare? - Black arrow on x-axis shows *actual* value - Black arrow on y-axis shows residual:<br> how *actual* value compares to distribution<br>of simulated values -- - Residuals are scaled (0 to 1) - If data fits model perfectly, expect all = 0.5 - Good fit *always* = flat/uniform distribution --- class: space-list # DHARMa Package ### *If* data fits the model ![:spacer 20px]() - Residuals follow a flat (uniform)<br>distribution (no matter what model!) -- - Expect: Straight line on QQ plot of<br>**uniform** distribution<br>.small[**(similar to QQ Normal plot)**] - Expect: No patterns between<br>residuals and model predictions<br>.small[**(similar to heteroscedasticity plot, resid vs. fitted)**] 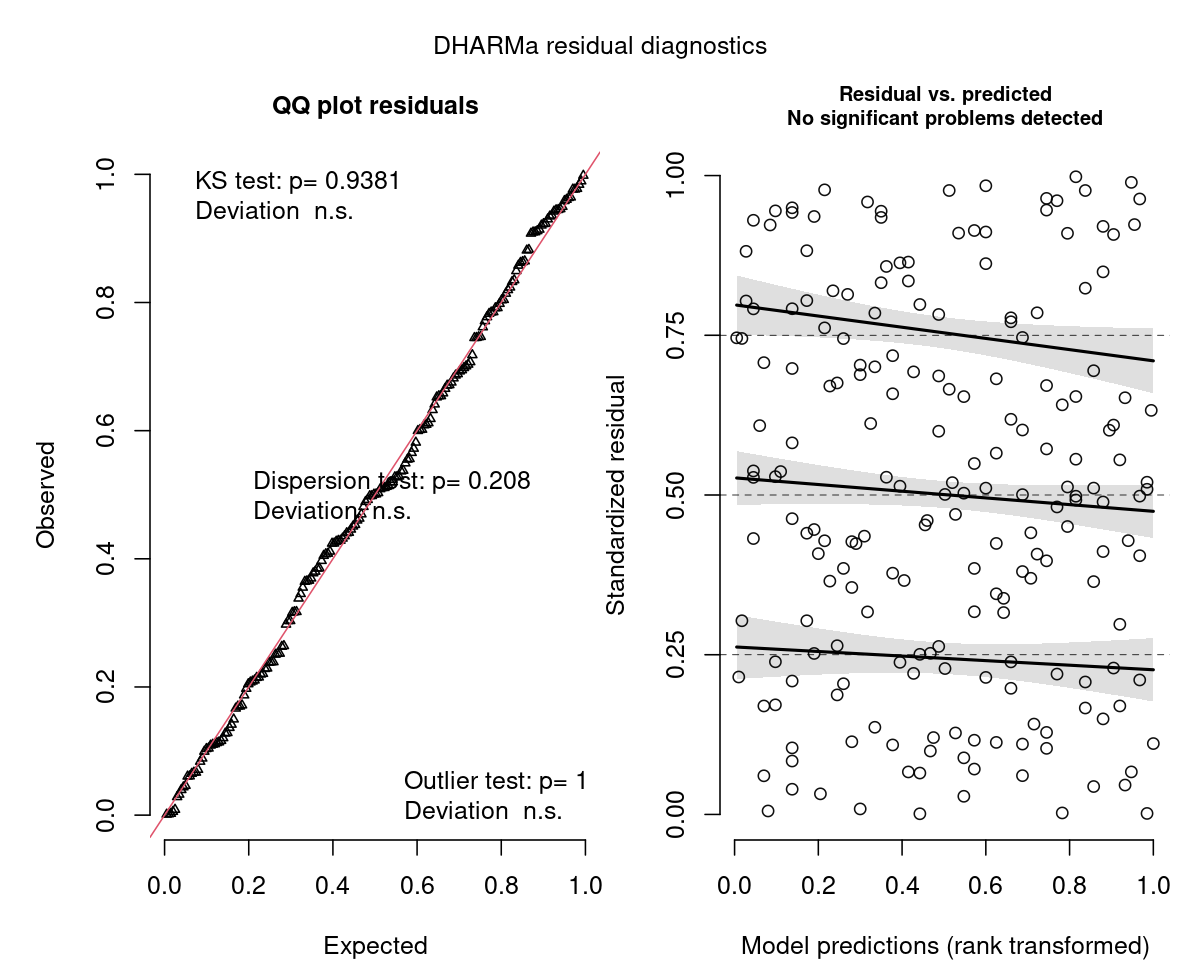 --  --- class: split-50 # DHARMa Package .columnl[ ### QQ Plot .medium[ - Tests **Uniformity** with Kolmogorov-Smirnov (KS) test - (do the residuals match a Uniform distribution?) - Tests for **Over/Underdispersion** with Dispersion Test - Tests for more **Outliers** than expected with Outlier test ] <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-1-1.png" width="100%" style="display: block; margin: auto;" /> ] -- .columnr[ ### Residuals vs. Predicted .medium[ - Check distribution of residuals .small[(visually and with quantiles)] - Dotted lines show expected quantiles - Black lines show simulated quantiles (want straight lines) - Outliers would show up as red stars ] <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-2-1.png" width="100%" style="display: block; margin: auto;" /> ] --- # DHARMa Package ### Usage ```r library(DHARMa) m <- lm(body_mass_g ~ flipper_length_mm, data = penguins) *r <- simulateResiduals(m, n = 1000, plot = TRUE) ``` - `simulateResiduals()` function from - Use `plot = TRUE` to produce diagnostic plots to see if simulated match expectation - `n = 1000` isn't strictly necessary but runs more simulations to produce more stable results -- ### Applies to any model - REMEMBER! Not assessing normality of residuals or heteroscedacity... - Assessing whether data fits the model - This *includes* assumptions, but *also* includes general fit, etc. - So, we can use this to see if our model could be improved... --- # DHARMa Package ### Check a previous model ```r m <- lm(body_mass_g ~ flipper_length_mm, data = penguins) r <- simulateResiduals(m, n = 1000, plot = TRUE) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-5-1.png" width="90%" style="display: block; margin: auto;" /> --  --- # DHARMa Package ### Improve the fit... <code class ='r hljs remark-code'>m <- lm(body_mass_g ~ flipper_length_mm + <span style="background-color:#ffff7f">species * sex</span>, data = penguins)<br>r <- simulateResiduals(m, n = 1000, plot = TRUE)</code> <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-7-1.png" width="90%" style="display: block; margin: auto;" /> --- class: section # Comparing Models ### Model Selection --- # Model Selection ### How do I know which model to use? 1. Check your diagnostics - Generally speaking, pick the model which has good diagnostics .spacer[] -- **All models have reasonably good diagnostics?** .spacer[] -- 2. Check model fit (how good is model at explaining data?) - Analysis of Variance/Deviance - `anova()` - Information Theoretic Approach (AIC) - `model.sel()` (MuMIn package) - (covered in class Nov 17th) -- > Many other methods, functions, packages --- # `anova()` - Analysis of Variance/Deviance ### Very common in R - Sequential analysis of variance for `lm()` - Analysis of deviance for `glm()` ```r p <- na.omit(penguins) # Must have exact same dataset for each model m1 <- lm(body_mass_g ~ flipper_length_mm, data = p) m2 <- lm(body_mass_g ~ flipper_length_mm + species + sex, data = p) m3 <- lm(body_mass_g ~ flipper_length_mm + species * sex, data = p) anova(m1, m2, m3) ``` --- # `anova()` - Analysis of Variance/Deviance ```r anova(m1, m2, m3) ``` ``` ## Analysis of Variance Table ## ## Model 1: body_mass_g ~ flipper_length_mm ## Model 2: body_mass_g ~ flipper_length_mm + species + sex ## Model 3: body_mass_g ~ flipper_length_mm + species * sex ## Res.Df RSS Df Sum of Sq F Pr(>F) ## 1 331 51211963 ## 2 328 28653568 3 22558395 91.082 < 2.2e-16 *** ## 3 326 26913579 2 1739989 10.538 3.675e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` --- # `anova()` - Analysis of Variance/Deviance ```r anova(m1, m2, m3) ``` ``` ## Analysis of Variance Table ## *## Model 1: body_mass_g ~ flipper_length_mm *## Model 2: body_mass_g ~ flipper_length_mm + species + sex *## Model 3: body_mass_g ~ flipper_length_mm + species * sex ## Res.Df RSS Df Sum of Sq F Pr(>F) ## 1 331 51211963 ## 2 328 28653568 3 22558395 91.082 < 2.2e-16 *** ## 3 326 26913579 2 1739989 10.538 3.675e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` - Models being compared --- # `anova()` - Analysis of Variance/Deviance ```r anova(m1, m2, m3) ``` ``` ## Analysis of Variance Table ## ## Model 1: body_mass_g ~ flipper_length_mm ## Model 2: body_mass_g ~ flipper_length_mm + species + sex ## Model 3: body_mass_g ~ flipper_length_mm + species * sex ## Res.Df RSS Df Sum of Sq F Pr(>F) *## 1 331 51211963 *## 2 328 28653568 3 22558395 91.082 < 2.2e-16 *** *## 3 326 26913579 2 1739989 10.538 3.675e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` - Models being compared - How the models compare --- # `anova()` - Analysis of Variance/Deviance ```r anova(m1, m2, m3) ``` ``` ## Analysis of Variance Table ## ## Model 1: body_mass_g ~ flipper_length_mm ## Model 2: body_mass_g ~ flipper_length_mm + species + sex ## Model 3: body_mass_g ~ flipper_length_mm + species * sex ## Res.Df RSS Df Sum of Sq F Pr(>F) ## 1 331 51211963 *## 2 328 28653568 3 22558395 91.082 < 2.2e-16 *** ## 3 326 26913579 2 1739989 10.538 3.675e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` - Models being compared - How the models compare - m2 has significantly (P < 0.05) lower Sums Squares than m1 --- # `anova()` - Analysis of Variance/Deviance ```r anova(m1, m2, m3) ``` ``` ## Analysis of Variance Table ## ## Model 1: body_mass_g ~ flipper_length_mm ## Model 2: body_mass_g ~ flipper_length_mm + species + sex ## Model 3: body_mass_g ~ flipper_length_mm + species * sex ## Res.Df RSS Df Sum of Sq F Pr(>F) ## 1 331 51211963 ## 2 328 28653568 3 22558395 91.082 < 2.2e-16 *** *## 3 326 26913579 2 1739989 10.538 3.675e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` - Models being compared - How the models compare - m2 has significantly (P < 0.05) lower Sums Squares than m1 - m3 has significantly (P < 0.05) lower Sums Squares than m2 --  --- # `model.sel()` - AIC model selection ```r library(MuMIn) model.sel(m1, m2, m3) ``` ``` ## Model selection table ## (Int) flp_lng_mm sex spc sex:spc family df logLik AICc delta weight ## m3 -478.3 20.49 + + + gaussian(identity) 8 -2353.956 4724.4 0.00 1 ## m2 -365.8 20.02 + + gaussian(identity) 6 -2364.387 4741.0 16.67 0 ## m1 -5872.0 50.15 gaussian(identity) 3 -2461.073 4928.2 203.86 0 ## Models ranked by AICc(x) ``` - m3 is the best (lowest AICc, and much lower than the others) - For more specifics on how to use/interpret this table, stay tuned for Nicky's lecture -- ![:spacer 20px]() > Always, remember that your 'best' model is only the best of what you've > compared... --- class: section # Generalized Linear Models [Chapter 13, "The R Book" by Michael J. Crawley](https://search.lib.umanitoba.ca/permalink/01UMB_INST/k6qbb2/cdi_skillsoft_books24x7_bks00051275) .small[(Freely available online through University of Manitoba Library)] --- # Generalized linear models  --  -- - Residuals are normally distributed - Normally distributed also known as "Gaussian" distribution --  --- # Generalized linear models - One type of Generalized linear model   -- - So what if your residuals are *not* normal? - ie. they don't follow a Gaussian distribution -- - Try a different family! --  --  --  -- - Etc. --- # Generalized linear models ### Gaussian Family .small[(Normal data)] ```r lm(y ~ x1 * x2, data = my_data) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-15-1.png" width="80%" style="display: block; margin: auto;" /> --- # Generalized linear models ### Poisson Family .small[(Count data)] ```r glm(counts ~ x1 * x2, family = "poisson", data = my_data) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-17-1.png" width="80%" style="display: block; margin: auto;" /> --- # Generalized linear models ### Negative Binomial Family .small[(Overdispersed Count data)] .small[ ```r MASS::glm.nb(counts ~ x1 * x2, data = my_data) ``` ] - Do Poisson model first - Check diagnostics, if necessary, you'll do a negative binomial model <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-19-1.png" width="80%" style="display: block; margin: auto;" /> --- # Generalized linear models ### Binomial Family .small[(Binary data)] - TRUE/FALSE, 0/1, Logistic Regression ```r glm(y ~ x1 * x2, family = "binomial", data = my_data) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-21-1.png" width="80%" style="display: block; margin: auto;" /> --- # Generalized linear models ### Binomial Family .small[(Binary data)] - TRUE/FALSE, 0/1, Logistic Regression ```r glm(y ~ x1 * x2, family = "binomial", data = my_data) ``` ### Binomial Family .small[(Proportion data)] - Two responses (binary outcomes), each in it's own column - .medium[(e.g., number of Yes, vs. No; Number of songs, vs. calls)] ```r glm(cbind(yes, no) ~ x1 * x2, family = "binomial", data = my_data) ``` --- class: section # Examples --- # Count Data - Poisson Family ### Get the data ```r p <- read_csv("https://stats.idre.ucla.edu/stat/data/poisson_sim.csv") p <- mutate(p, program = factor(prog, levels = 1:3, labels = c("General", "Academic", "Vocational"))) p ``` .small[ ``` ## # A tibble: 200 × 5 ## id num_awards prog math program ## <dbl> <dbl> <dbl> <dbl> <fct> ## 1 45 0 3 41 Vocational ## 2 108 0 1 41 General ## 3 15 0 3 44 Vocational ## 4 67 0 3 42 Vocational ## 5 153 0 3 40 Vocational ## 6 51 0 1 42 General ## 7 164 0 3 46 Vocational ## 8 133 0 3 40 Vocational ## 9 2 0 3 33 Vocational ## 10 53 0 3 46 Vocational ## # … with 190 more rows ``` ] ?) --- # Count Data - Poisson Family ### Look at the data ```r ggplot(data = p, aes(x = math, y = num_awards, colour = program)) + geom_point() ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-25-1.png" width="75%" style="display: block; margin: auto;" /> --- # Count Data - Poisson Family ### Look at the data ```r ggplot(data = p, aes(x = math, y = num_awards, colour = program)) + geom_point() + * stat_smooth(method = "glm", se = FALSE) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-26-1.png" width="70%" style="display: block; margin: auto;" /> --- # Count Data - Poisson Family ### Look at the data ```r ggplot(data = p, aes(x = math, y = num_awards, colour = program)) + geom_point() + * stat_smooth(method = "glm", method.args = list(family = "poisson"), se = FALSE) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-27-1.png" width="70%" style="display: block; margin: auto;" /> --- class: split-60 # Count Data - Poisson Family ### Run the model ```r m <- glm(num_awards ~ math + program, family = "poisson", data = p) ``` --- class: split-60 # Count Data - Poisson Family .columnl[ ### Diagnostics ```r summary(m) ``` .small[ ``` ## Call: ## glm(formula = num_awards ~ math + program, family = "poisson", ## data = p) ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -5.24712 0.65845 -7.969 1.60e-15 *** ## math 0.07015 0.01060 6.619 3.63e-11 *** ## programAcademic 1.08386 0.35825 3.025 0.00248 ** ## programVocational 0.36981 0.44107 0.838 0.40179 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## *## (Dispersion parameter for poisson family taken to be 1) ## ## Null deviance: 287.67 on 199 degrees of freedom ## Residual deviance: 189.45 on 196 degrees of freedom ## AIC: 373.5 ## ## Number of Fisher Scoring iterations: 6 ``` ] ] .columnr[ ### Overdispersion .medium[ - Dispersion = Residual Deviance / DF - Expected to be 1 for Poisson - Overdispersion > 1; Underdispersion < 1 ]] --- class: split-60 # Count Data - Poisson Family .columnl[ ### Diagnostics ```r summary(m) ``` .small[ ``` ## Call: ## glm(formula = num_awards ~ math + program, family = "poisson", ## data = p) ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -5.24712 0.65845 -7.969 1.60e-15 *** ## math 0.07015 0.01060 6.619 3.63e-11 *** ## programAcademic 1.08386 0.35825 3.025 0.00248 ** ## programVocational 0.36981 0.44107 0.838 0.40179 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for poisson family taken to be 1) ## ## Null deviance: 287.67 on 199 degrees of freedom *## Residual deviance: 189.45 on 196 degrees of freedom ## AIC: 373.5 ## ## Number of Fisher Scoring iterations: 6 ``` ] ] .columnr[ ### Overdispersion .medium[ - Dispersion = Residual Deviance / DF - Expected to be 1 for Poisson - Overdispersion > 1; Underdispersion < 1 ] .medium[ **Traditional check:** - Residual Deviance (189.45) vs. DF (196) ] .small[ ```r deviance(m) / df.residual(m) ``` ``` ## [1] 0.9665797 ``` ] .medium[ - Almost 1 .small[(which it is good)] ] .small[ ```r pchisq(deviance(m), df.residual(m), lower.tail = FALSE) ``` ``` ## [1] 0.6182274 ``` ] .medium[ - Test shows no significant overdispersion (p = 0.62) ]] --- class: split-50 # Count Data - Poisson Family ### Diagnostics - Residuals are complicated to assess in GLMs - Therefore, use `DHARMa` package! ```r r <- simulateResiduals(m, n = 1000, plot = TRUE) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/poisson-1.png" width="100%" style="display: block; margin: auto;" /> -- </small>) --- class: split-50 # Count Data - Poisson Family - Model Diagnostics ### Zero-inflation .small[(More zeros than expected)] - Will often pop up as iffy residuals - Overdispersion can lead to false positives - Here not a problem (non-significant P-value) ```r testZeroInflation(m, plot = FALSE) ``` ``` ## ## DHARMa zero-inflation test via comparison to expected zeros with simulation under H0 = ## fitted model ## ## data: simulationOutput ## ratioObsSim = 1.0131, p-value = 0.832 ## alternative hypothesis: two.sided ``` --- class: split-60, space-list # Count Data - Poisson Family .columnl[ ### Interpretation ```r summary(m) ``` .small[ ``` ## Call: ## glm(formula = num_awards ~ math + program, family = "poisson", ## data = p) ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) *## (Intercept) -5.24712 0.65845 -7.969 1.60e-15 *** *## math 0.07015 0.01060 6.619 3.63e-11 *** *## programAcademic 1.08386 0.35825 3.025 0.00248 ** *## programVocational 0.36981 0.44107 0.838 0.40179 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for poisson family taken to be 1) ## ## Null deviance: 287.67 on 199 degrees of freedom ## Residual deviance: 189.45 on 196 degrees of freedom ## AIC: 373.5 ## ## Number of Fisher Scoring iterations: 6 ``` ] ] .columnr[ ### Effects .medium[ - Significantly more awards received with higher marks in math - i.e. Number of awards increases by 0.07 log-counts per 1 unit increase in Math mark - Significantly more awards received in Academic Program compared to General - i.e. Number of awards greater by 1.08 log-counts for Academic compared to General - No difference in amount of awards received in Vocational vs. General Program ]] --  --- class: split-60, space-list # Count Data - Poisson Family .columnl[ ### Interpretation ```r summary(m) ``` .small[ ``` ## Call: ## glm(formula = num_awards ~ math + program, family = "poisson", ## data = p) ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) *## (Intercept) -5.24712 0.65845 -7.969 1.60e-15 *** *## math 0.07015 0.01060 6.619 3.63e-11 *** *## programAcademic 1.08386 0.35825 3.025 0.00248 ** *## programVocational 0.36981 0.44107 0.838 0.40179 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for poisson family taken to be 1) ## ## Null deviance: 287.67 on 199 degrees of freedom ## Residual deviance: 189.45 on 196 degrees of freedom ## AIC: 373.5 ## ## Number of Fisher Scoring iterations: 6 ``` ] ] .columnr[ ### Interpreting Results .medium[ - Convert to ratios with `\(e^{\textrm{est}}\)` (`exp()`) ] .small[ ```r data.frame(est = coef(m), ratios = exp(coef(m))) ``` ``` ## est ratios ## (Intercept) -5.2471244 0.00526263 ## math 0.0701524 1.07267164 ## programAcademic 1.0838591 2.95606545 ## programVocational 0.3698092 1.44745846 ``` ] .medium[ - No. awards increases by 1.07 *times* per 1 unit increase in Math mark (7%) - No. awards received by Academic is 2.96 *times* greater than in General program ]] --- # Overdispersion .medium[ - Overdispersion when data is spread out more than distribution would be (longer tails) - Results in **highly** significant findings that are **not** valid!! - Simulated residuals run from 0 to 1, but here more residuals around 0 and 1 (longer tails) ] <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/overdisp-1.png" width="100%" style="display: block; margin: auto;" /> --- # Underdispersion .medium[ - Underdispersion is less common - When data is gathered towards the centre more than distribution would be (shorter tails) - These simulated residuals run from 0 to 1 - Here, more residuals around 0.5 (shorter tails) ] <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/underdisp-1.png" width="100%" style="display: block; margin: auto;" /> --- class: split-50 # Overdispersed Count Data - Negative Binomial ```r quine <- MASS::quine ``` .columnl[ ### Poisson GLM .medium[ <code class ='r hljs remark-code'>m1 <- <span style="background-color:#ffff7f">glm</span>(Days ~ Sex, data = quine, <span style="background-color:#ffff7f">family = "poisson"</span>)<br>summary(m1)</code> ] .small[ ``` ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 2.72294 0.02865 95.030 < 2e-16 *** ## SexM 0.16490 0.04080 4.041 5.31e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for poisson family taken to be 1) ## ## Null deviance: 2073.5 on 145 degrees of freedom ## Residual deviance: 2057.2 on 144 degrees of freedom ``` ]] .columnr[ ### Negative Binomial GLM .medium[ <code class ='r hljs remark-code'>m2 <- <span style="background-color:#ffff7f">MASS::glm.nb</span>(Days ~ Sex, data = quine)<br>summary(m2)</code> ] .small[ ``` ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 2.7229 0.1116 24.395 <2e-16 *** ## SexM 0.1649 0.1656 0.996 0.319 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for Negative Binomial(1.0741) family taken to be 1) ## ## Null deviance: 169.50 on 145 degrees of freedom ## Residual deviance: 168.51 on 144 degrees of freedom ``` ]] --- class: split-50 # Overdispersed Count Data - Negative Binomial ```r quine <- MASS::quine ``` .columnl[ ### Poisson GLM .medium[ <code class ='r hljs remark-code'>m1 <- <span style="background-color:#ffff7f">glm</span>(Days ~ Sex, data = quine, <span style="background-color:#ffff7f">family = "poisson"</span>)<br>summary(m1)</code> ] .small[ ``` ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 2.72294 0.02865 95.030 < 2e-16 *** *## SexM 0.16490 0.04080 4.041 5.31e-05 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for poisson family taken to be 1) ## ## Null deviance: 2073.5 on 145 degrees of freedom *## Residual deviance: 2057.2 on 144 degrees of freedom ``` ]] .columnr[ ### Negative Binomial GLM .medium[ <code class ='r hljs remark-code'>m2 <- <span style="background-color:#ffff7f">MASS::glm.nb</span>(Days ~ Sex, data = quine)<br>summary(m2)</code> ] .small[ ``` ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 2.7229 0.1116 24.395 <2e-16 *** *## SexM 0.1649 0.1656 0.996 0.319 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for Negative Binomial(1.0741) family taken to be 1) ## ## Null deviance: 169.50 on 145 degrees of freedom *## Residual deviance: 168.51 on 144 degrees of freedom ``` ]] --- class: split-50 # Overdispersed Count Data - Negative Binomial .columnl[ ### Poisson GLM ```r r <- simulateResiduals(m1) plotQQunif(r) # Just the first Uniformity Plot ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-51-1.png" width="100%" style="display: block; margin: auto;" /> ] .columnr[ ### Negative Binomial GLM ```r r <- simulateResiduals(m2) plotQQunif(r) # Just the first Uniformity Plot ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-52-1.png" width="100%" style="display: block; margin: auto;" /> ] --  --- # Binary Data (0/1) - Binomial Family .small[(logistic regresion)] ### Get the data - Mountain chickadees atypical songs by urbanization - Negative `urbanization` more rural - Positive `urbanization` more urban - `atypical_c` atypical singer (1) or 'normal' singer (0) ```r atypical <- read_csv("https://steffilazerte.ca/NRI_7350/data/atypical.csv") atypical ``` ``` ## # A tibble: 78 × 2 ## atypical urbanization ## <dbl> <dbl> ## 1 1 0.540 ## 2 0 0.582 ## 3 0 -0.950 ## 4 0 -0.950 ## 5 0 0.513 ## 6 0 1.69 ## 7 0 1.51 ## 8 0 1.72 ## 9 0 1.28 ## 10 0 1.43 ## # … with 68 more rows ```  --- # Binary Data (0/1) - Binomial Family .small[(logistic regresion)] ### Look at the data ```r ggplot(data = atypical, aes(x = urbanization, y = atypical)) + geom_point() + stat_smooth(method = "glm", method.args = list(family = "binomial")) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-55-1.png" width="70%" style="display: block; margin: auto;" />  --- # Binary Data (0/1) - Binomial Family .small[(logistic regresion)] ### Run model and check diagnostics .small[ ```r m <- glm(atypical ~ urbanization, family = "binomial", data = atypical) r <- simulateResiduals(m, plot = TRUE) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-56-1.png" width="100%" style="display: block; margin: auto;" /> ]  --- class: split-60 # Binary Data (0/1) - Binomial Family .small[(logistic regresion)] .columnl[ ```r summary(m) ``` .small[ ``` ## ## Call: ## glm(formula = atypical ~ urbanization, family = "binomial", data = atypical) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -0.9907 -0.4460 -0.2500 -0.2210 2.7201 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -2.6572 0.5380 -4.939 7.85e-07 *** ## urbanization 1.1000 0.4209 2.613 0.00897 ** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## ## Null deviance: 51.586 on 77 degrees of freedom ## Residual deviance: 43.203 on 76 degrees of freedom ## AIC: 47.203 ## ## Number of Fisher Scoring iterations: 6 ``` ] ] .columnr[ ### Interpreting Results ```r exp(coef(m)) ``` ``` ## (Intercept) urbanization ## 0.07014254 3.00406665 ``` .medium[ E.g., The odds of being an atypical singer increase by a factor of 3 (x3 times more likely) for every unit increase in Habit Urbanization. ]]  --- # Binary Outcomes - Binomial Family Proportion with binary outcomes (e.g., 10 yes, 5 no) ### Get the data ```r admissions <- as.data.frame(UCBAdmissions) admissions <- pivot_wider(admissions, names_from = Admit, values_from = Freq) admissions ``` ``` ## # A tibble: 12 × 4 ## Gender Dept Admitted Rejected ## <fct> <fct> <dbl> <dbl> ## 1 Male A 512 313 ## 2 Female A 89 19 ## 3 Male B 353 207 ## 4 Female B 17 8 ## 5 Male C 120 205 ## 6 Female C 202 391 ## 7 Male D 138 279 ## 8 Female D 131 244 ## 9 Male E 53 138 ## 10 Female E 94 299 ## 11 Male F 22 351 ## 12 Female F 24 317 ``` --- class: split-60 # Binary Outcomes - Binomial Family .columnl[ ### Diagnostics ```r m <- glm(cbind(Admitted, Rejected) ~ Gender, family = "binomial", data = admissions) summary(m) ``` .small[ ``` ## Call: ## glm(formula = cbind(Admitted, Rejected) ~ Gender, family = "binomial", ## data = admissions) ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.22013 0.03879 -5.675 1.38e-08 *** ## GenderFemale -0.61035 0.06389 -9.553 < 2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## *## Null deviance: 877.06 on 11 degrees of freedom ## Residual deviance: 783.61 on 10 degrees of freedom ## AIC: 856.55 ## ## Number of Fisher Scoring iterations: 4 ``` ] ] .columnr[ ### Overdispersion .medium[ **Traditional check:** - Look at resid deviance (783.61) vs. df (10) ] .small[ ```r deviance(m) / df.residual(m) ``` ``` ## [1] 78.3607 ``` ] .medium[ - Very large .small[(definitely not close to 1)] ] .small[ ```r pchisq(deviance(m), df.residual(m), lower.tail = FALSE) ``` ``` ## [1] 6.892992e-162 ``` ] .medium[ - Test shows significant overdispersion ]] --- # Binary Outcomes - Binomial Family ### Check with DHARMa ```r plotQQunif(m) plotResiduals(m, asFactor = TRUE) # to ensure Gender is treated as category ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-65-1.png" width="100%" style="display: block; margin: auto;" />  --- # Binary Outcomes - Quasi-binomial Family .small[for overdispersion] ```r m_quasi <- glm(cbind(Admitted, Rejected) ~ Gender, family = "quasibinomial", data = admissions) summary(m_quasi) ``` .small[ ``` ## ## Call: ## glm(formula = cbind(Admitted, Rejected) ~ Gender, family = "quasibinomial", ## data = admissions) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -16.7915 -4.7613 -0.4365 5.1025 11.2022 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -0.2201 0.3281 -0.671 0.517 ## GenderFemale -0.6104 0.5404 -1.129 0.285 ## *## (Dispersion parameter for quasibinomial family taken to be 71.52958) ## ## Null deviance: 877.06 on 11 degrees of freedom ## Residual deviance: 783.61 on 10 degrees of freedom ## AIC: NA ## ## Number of Fisher Scoring iterations: 4 ``` ] --  --- # Binary Outcomes - Mixed models .small[for overdispersion] - Observation-level random effects (i.e assign each observation an ID number) ```r library(lme4) admissions <- mutate(admissions, ID = 1:n()) m_glmm <- glmer(cbind(Admitted, Rejected) ~ Gender + (1|ID), family = "binomial", data = admissions) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-67-1.png" width="100%" style="display: block; margin: auto;" /> --- class: split-50 # Binary Outcomes - Overdispersion Either way, fixing overdispersion remove the 'significance' ### Original ```r coef(summary(m)) ``` .small[ ``` ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.2201340 0.03878810 -5.675297 1.384479e-08 *## GenderFemale -0.6103524 0.06389305 -9.552720 1.263352e-21 ``` ] ![:spacer 10px]() .columnl[ ### Quasi fix ```r coef(summary(m_quasi)) ``` .small[ ``` ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -0.2201340 0.3280510 -0.6710359 0.5173966 *## GenderFemale -0.6103524 0.5403765 -1.1294947 0.2850545 ``` ] ] .columnr[ ### GLMM fix ```r coef(summary(m_glmm)) ``` .small[ ``` ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.6508845 0.4932596 -1.319558 0.1869827 *## GenderFemale 0.1747094 0.7016048 0.249014 0.8033499 ``` ] ] --- class: split-30 # Your Turn! .columnl[ We have the `crabs` dataset ] .columnr[ ```r crabs <- read_csv("https://steffilazerte.ca/NRI_7350/data/crabs.csv") ``` ] ![:spacer 20px]() ### Background - Horseshoe crabs form pairs for spawning (mating) - But extra, unattached males crowd around and try to get involved (Satellite males) ### Your Job You're interested in the effect of female size (`width`) on the number of male `satellites` - Look at your data (make a plot) - Run a `glm()` for count data - Check your diagnostics. Do you have a problem? Check for overdispersion and zero-inflation - Apply an overdispersion fix - Check your diagnostics. Do you have a problem? Check for zero-inflation  .footnote[Brockmann HJ (1996). “Satellite Male Groups in Horseshoe Crabs, *Limulus polyphemus*”, Ethology, 102(1), 1–21.] --- exclude: FALSE # Your Turn!  ### Look at your data ```r ggplot(data = crabs, aes(x = weight, y = satellites)) + geom_point() + stat_smooth(method = "glm", method.args = list(family = "poisson")) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-73-1.png" width="70%" style="display: block; margin: auto;" /> --- exclude: FALSE # Your Turn!  ### Run a `glm()` for count data .small[ ```r m <- glm(satellites ~ weight, data = crabs, family = "poisson") summary(m) ``` ``` ## ## Call: ## glm(formula = satellites ~ weight, family = "poisson", data = crabs) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -2.9306 -1.9981 -0.5627 0.9299 4.9992 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.4282 0.1789 -2.394 0.0167 * ## weight 0.5892 0.0650 9.065 <2e-16 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for poisson family taken to be 1) ## ## Null deviance: 632.79 on 172 degrees of freedom ## Residual deviance: 560.84 on 171 degrees of freedom ## AIC: 920.14 ## ## Number of Fisher Scoring iterations: 5 ``` ] --- exclude: FALSE # Your Turn!  ### Check your diagnostics - Do you have a problem? Check for overdispersion and zero-inflation ```r r <- simulateResiduals(m, plot = TRUE) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-75-1.png" width="100%" style="display: block; margin: auto;" /> --- class: split-50 exclude: FALSE # Your Turn!  ### Check your diagnostics - Do you have a problem? Check for overdispersion and zero-inflation ```r testZeroInflation(m) ``` .columnl[ .small[ ``` ## ## DHARMa zero-inflation test via comparison to expected zeros with simulation under H0 = ## fitted model ## ## data: simulationOutput ## ratioObsSim = 4.381, p-value < 2.2e-16 ## alternative hypothesis: two.sided ``` ]] .columnr[ <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-77-1.png" width="100%" style="display: block; margin: auto;" /> ] --- exclude: FALSE # Your Turn!  ### Apply an overdispersion fix .small[ ```r m <- MASS::glm.nb(satellites ~ weight, data = crabs) summary(m) ``` ``` ## ## Call: ## MASS::glm.nb(formula = satellites ~ weight, data = crabs, init.theta = 0.9310998551, ## link = log) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -1.8393 -1.4123 -0.3246 0.4747 2.1278 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -0.8637 0.4046 -2.135 0.0328 * ## weight 0.7599 0.1578 4.817 1.46e-06 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for Negative Binomial(0.9311) family taken to be 1) ## ## Null deviance: 216.44 on 172 degrees of freedom ## Residual deviance: 196.16 on 171 degrees of freedom ## AIC: 754.64 ## ## Number of Fisher Scoring iterations: 1 ## ## ## Theta: 0.931 ## Std. Err.: 0.168 ## ## 2 x log-likelihood: -748.643 ``` ] --- exclude: FALSE # Your Turn!  ### Check your diagnostics - Do you still have a problem? Check for overdispersion and zero-inflation ```r r <- simulateResiduals(m, plot = TRUE) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-79-1.png" width="100%" style="display: block; margin: auto;" /> --- class: split-50 exclude: FALSE # Your Turn!  ### Check your diagnostics - Do you still have a problem? Check for overdispersion and zero-inflation ```r testZeroInflation(m) ``` .columnl[ .small[ ``` ## ## DHARMa zero-inflation test via comparison to expected zeros with simulation under H0 = ## fitted model ## ## data: simulationOutput ## ratioObsSim = 1.2358, p-value = 0.088 ## alternative hypothesis: two.sided ``` ]] .columnr[ <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-81-1.png" width="100%" style="display: block; margin: auto;" /> ]  --- # Zero-inflated Models .small[(Advanced example! Above and beyond!)]  - `glmmTMB()` function from `glmmTMB` package - Allows modeling zero-inflation (`ziformula` says zeros are a function of the variable `weight`) .small[ ```r crabs <- read_csv("https://steffilazerte.ca/NRI_7350/data/crabs.csv") library(glmmTMB) m <- glmmTMB(satellites ~ weight, ziformula = ~ weight , family = "nbinom2", data = crabs) summary(m) ``` ``` ## Dispersion parameter for nbinom2 family (): 4.96 ## ## Conditional model: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.8979 0.3051 2.943 0.00325 ** ## weight 0.2171 0.1118 1.942 0.05217 . ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Zero-inflation model: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 3.7546 0.9837 3.817 0.000135 *** ## weight -1.9123 0.4320 -4.426 9.59e-06 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ] --- # Zero-inflated Models .small[(Advanced example! Above and beyond!)]  - `glmmTMB()` function from `glmmTMB` package - Allows modeling zero-inflation (`ziformula` says zeros are a function of the variable `weight`) .small[ ```r crabs <- read_csv("https://steffilazerte.ca/NRI_7350/data/crabs.csv") library(glmmTMB) m <- glmmTMB(satellites ~ weight, ziformula = ~ weight , family = "nbinom2", data = crabs) summary(m) ``` ``` ## Dispersion parameter for nbinom2 family (): 4.96 ## *## Conditional model: *## Estimate Std. Error z value Pr(>|z|) *## (Intercept) 0.8979 0.3051 2.943 0.00325 ** *## weight 0.2171 0.1118 1.942 0.05217 . ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Zero-inflation model: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 3.7546 0.9837 3.817 0.000135 *** ## weight -1.9123 0.4320 -4.426 9.59e-06 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ] ) --- # Zero-inflated Models .small[(Advanced example! Above and beyond!)]  - `glmmTMB()` function from `glmmTMB` package - Allows modeling zero-inflation (`ziformula` says zeros are a function of the variable `weight`) .small[ ```r crabs <- read_csv("https://steffilazerte.ca/NRI_7350/data/crabs.csv") library(glmmTMB) m <- glmmTMB(satellites ~ weight, ziformula = ~ weight , family = "nbinom2", data = crabs) summary(m) ``` ``` ## Dispersion parameter for nbinom2 family (): 4.96 ## ## Conditional model: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) 0.8979 0.3051 2.943 0.00325 ** ## weight 0.2171 0.1118 1.942 0.05217 . ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## *## Zero-inflation model: *## Estimate Std. Error z value Pr(>|z|) *## (Intercept) 3.7546 0.9837 3.817 0.000135 *** *## weight -1.9123 0.4320 -4.426 9.59e-06 *** ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` ]  --- # Zero-inflated Models .small[(Advanced example! Above and beyond!)] ```r r <- simulateResiduals(m, plot = TRUE) ``` <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-88-1.png" width="100%" style="display: block; margin: auto;" />  .footnote[For more information on the glmmTMB package see their [Journal Article](https://journal.r-project.org/archive/2017/RJ-2017-066/RJ-2017-066.pdf)] --- class: split-50 # Zero-inflated Models .small[(Advanced example! Above and beyond!)]  .footnote[For more information on the glmmTMB package see their [Journal Article](https://journal.r-project.org/archive/2017/RJ-2017-066/RJ-2017-066.pdf)] ```r testZeroInflation(m) ``` .columnl[ .small[ ``` ## ## DHARMa zero-inflation test via comparison to expected zeros with simulation under H0 = ## fitted model ## ## data: simulationOutput ## ratioObsSim = 1.0016, p-value = 1 ## alternative hypothesis: two.sided ``` ]] .columnr[ <img src="6 GLMs and Non-parametric stats - answers_files/figure-html/unnamed-chunk-90-1.png" width="100%" style="display: block; margin: auto;" /> ] --- class: section # Packages and References for <p> Other Advanced Models --- # Packages and References ### (Generalized) Linear Mixed Models (LMM, GLMM) - Also called generalized linear mixed effects models (GLME, LME) - `lme4` - More advanced, crossed-random factors, Generalized (`glmer()`) and Gaussian (`lmer()`) - `nlme` - Older but can specify auto-correlation structures, only Gaussian (`lme()`) - `glmmTMB` - Zero-inflated models and other distribtions **References** - [Ben Bolker's GLMM FAQ](https://bbolker.github.io/mixedmodels-misc/glmmFAQ.html) - [Chapter 19, "The R Book" by Michael J. Crawley](https://search.lib.umanitoba.ca/permalink/01UMB_INST/k6qbb2/cdi_skillsoft_books24x7_bks00051275) .small[(Freely available online through University of Manitoba Library)] - [Mixed Effects Models and Extensions in Ecology with R by Alain Zuur](https://search.lib.umanitoba.ca/permalink/01UMB_INST/1p55dqn/alma99149017057101651) .small[(Freely available online through University of Manitoba Library)] - [Generalized linear mixed models: a practical guide for ecology and evolution](https://search.lib.umanitoba.ca/permalink/01UMB_INST/k6qbb2/cdi_proquest_miscellaneous_66983023), 2009, Trends in ecology and evolution --- # Packages and References ### General Additive Models (GAM) - `mgcv` package (`gam()`, `gamm()`) - `gamm4` package (`gamm4()`) **References** - [Chapter 19, "The R Book" by Michael J. Crawley](https://search.lib.umanitoba.ca/permalink/01UMB_INST/k6qbb2/cdi_skillsoft_books24x7_bks00051275) .small[(Freely available online through University of Manitoba Library)] - "Generalized Additive Models: An Introduction with R" by Simon N. Wood .small[(Hard-copy available from University of Manitoba Library)] --- # Packages and References ### Generalized Estimating Equations (GEE) - `gee` package (`gee()`) - `geepack` package (`geeglm()`) **References** - [The R package geepack for Generalized Estimating equations](https://www.jstatsoft.org/article/view/v015i02), Journal of Statistical Software, 2005 - [geepack Manual](https://cran.r-project.org/web/packages/geepack/vignettes/geepack-manual.pdf) --- class: section # Non-parametric Statistics --- # Non-parametric Statistics ### Wilcoxon Rank Sum (Mann-Whitney) Test ```r air <- filter(airquality, Month %in% c(5, 8)) ``` Is there a difference in air quality between May (5th month) and August (8th month)? ```r wilcox.test(Ozone ~ Month, data = air, exact = FALSE) ``` ``` ## ## Wilcoxon rank sum test with continuity correction ## ## data: Ozone by Month ## W = 127.5, p-value = 0.0001208 ## alternative hypothesis: true location shift is not equal to 0 ``` Yes! --- # Non-parametric Statistics ### Kruskal-Wallis Rank Sum Test Is there a difference in air quality among months? ```r kruskal.test(Ozone ~ Month, data = airquality) ``` ``` ## ## Kruskal-Wallis rank sum test ## ## data: Ozone by Month ## Kruskal-Wallis chi-squared = 29.267, df = 4, p-value = 6.901e-06 ``` Yes, there is at least one month that is different from the rest.