source("XX_functions.R") # Custom functions and packages
# Metrics
v <- read_csv("Data/Datasets/vultures_final.csv") |>
# Round non-integer values of population counts
mutate(across(c(contains("pop"), contains("raw")), round))
# Raw counts
raw <- read_csv("Data/Datasets/vultures_clean_2023.csv")
# Predicted GAM models
pred <- read_csv("Data/Datasets/vultures_gams_pred.csv")
# Checking problematic years
supp <- read_csv("Data/Datasets/table_supplemental.csv")
# Problematic years
v <- mutate(v, gaps = if_else(year %in% c(2011, 2013), "Gap around peak", "No gaps"))
y1 <- c(2011, 2013) # Gaps around the peak
y2 <- c(2001, 2006, 2023) # Missing end of migrationManuscript Figure and Supplemental
This is the final figure and Supplemental material for the manuscript.
Setup
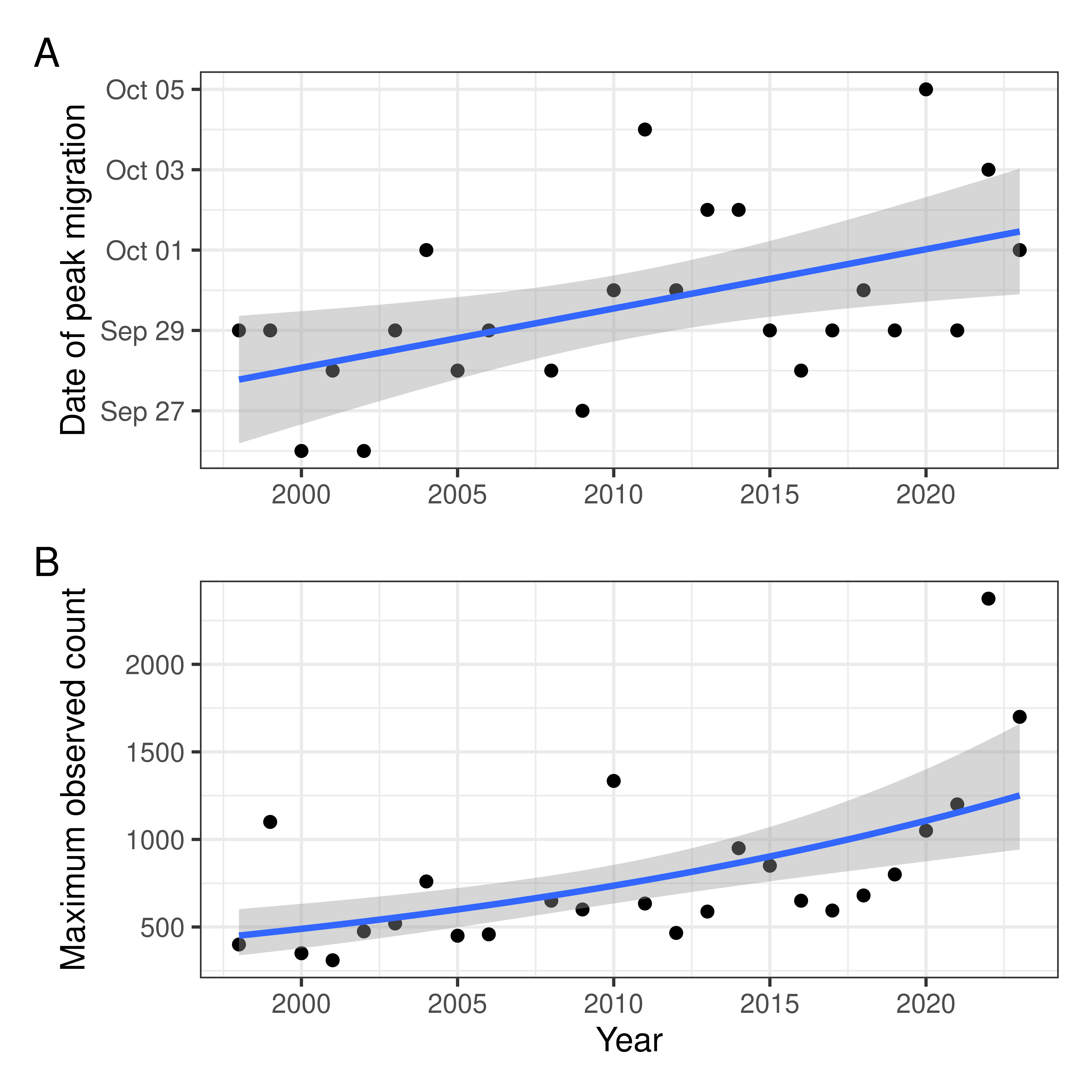
Main figure
Code
v <- v |>
mutate(date = as_date(p50_doy) - days(1))
g1 <- ggplot(v, aes(x = year, y = date)) +
theme_bw() +
theme(axis.title.x = element_blank(),
legend.direction = "vertical", legend.position = "inside",
legend.position.inside = c(0.1, 0.85),
legend.key.width = unit(2, "pt"),
legend.key.spacing.y = unit(-5, "pt"),
legend.background = element_blank(),
legend.key = element_blank(),
legend.margin = margin(-10, 0, 0, 0, "pt"),
plot.margin = margin(1,2,0,1, "pt"), panel.spacing = unit(0, "pt")) +
geom_point(aes(shape = gaps)) +
geom_smooth(method = "lm", se = TRUE) +
scale_shape_manual(name = NULL, values = c(1, 19)) +
labs(y = "Date of Peak Migration", x = "")
g2 <- ggplot(v, aes(x = year, y = mig_raw_max)) +
theme_bw() +
theme(legend.position = "none") +
geom_point(aes(shape = gaps)) +
stat_smooth(method = MASS::glm.nb) +
scale_shape_manual(name = NULL, values = c(1, 19)) +
labs(y = "Annual Maximum\nDaily Estimated Total", x = "Year")
g3 <- ggplot(v, aes(x = year, y = mig_raw_max)) +
theme_bw() +
theme(legend.direction = "horizontal", legend.position = "inside",
legend.position.inside = c(0.25, 0.87),
legend.background = element_blank(),
legend.key = element_blank(),
legend.margin = margin(-10, 0, 0, 0, "pt"),
plot.margin = margin(1,2,0,1, "pt"), panel.spacing = unit(0, "pt")) +
geom_point() +
stat_smooth(method = MASS::glm.nb, aes(fill = "All years"), alpha = 0.15) +
stat_smooth(data = filter(v, year != 2022), aes(fill = "Without 2022"),
method = MASS::glm.nb, linewidth = 1, colour = "black") +
labs(y = "Annual Maximum\nDaily Estimated Total", x = "Year") +
scale_fill_manual(name = NULL,
values = c("All years" = "blue", "Without 2022" = "darkgrey"))
g <- g1 / g2 +
plot_annotation(tag_levels = "A") #+
# plot_layout(guides = "collect") &
# theme(legend.direction = "horizontal", legend.position = "bottom",
# legend.key.width = unit(2, "pt"),
# legend.background = element_blank(),
# legend.key = element_blank(),
# legend.margin = margin(-10, 0, 0, 0, "pt"),
# plot.margin = margin(1,2,0,1, "pt"), panel.spacing = unit(0, "pt"))
#ggsave("fig1_quick.png", dpi = 1000, width = 8, height = 7)
gg <- g1 / (g2 + labs(y = "Annual Maximum DET")) + plot_annotation(tag_levels = "A")Double checking growth and increases
Code
m <- MASS::glm.nb(mig_raw_max ~ year, data = v)
d <- select(v, year) |>
mutate(y = predict(m, v),
yexp = exp(y))
# Compound growth (avg growth over the period)
(1251 / 451)^ (1/25) - 1[1] 0.04165339Code
# Compound interest (amount at the end)
451 * (1 + 0.041647)^(25)[1] 1250.808Code
# Factor of increase
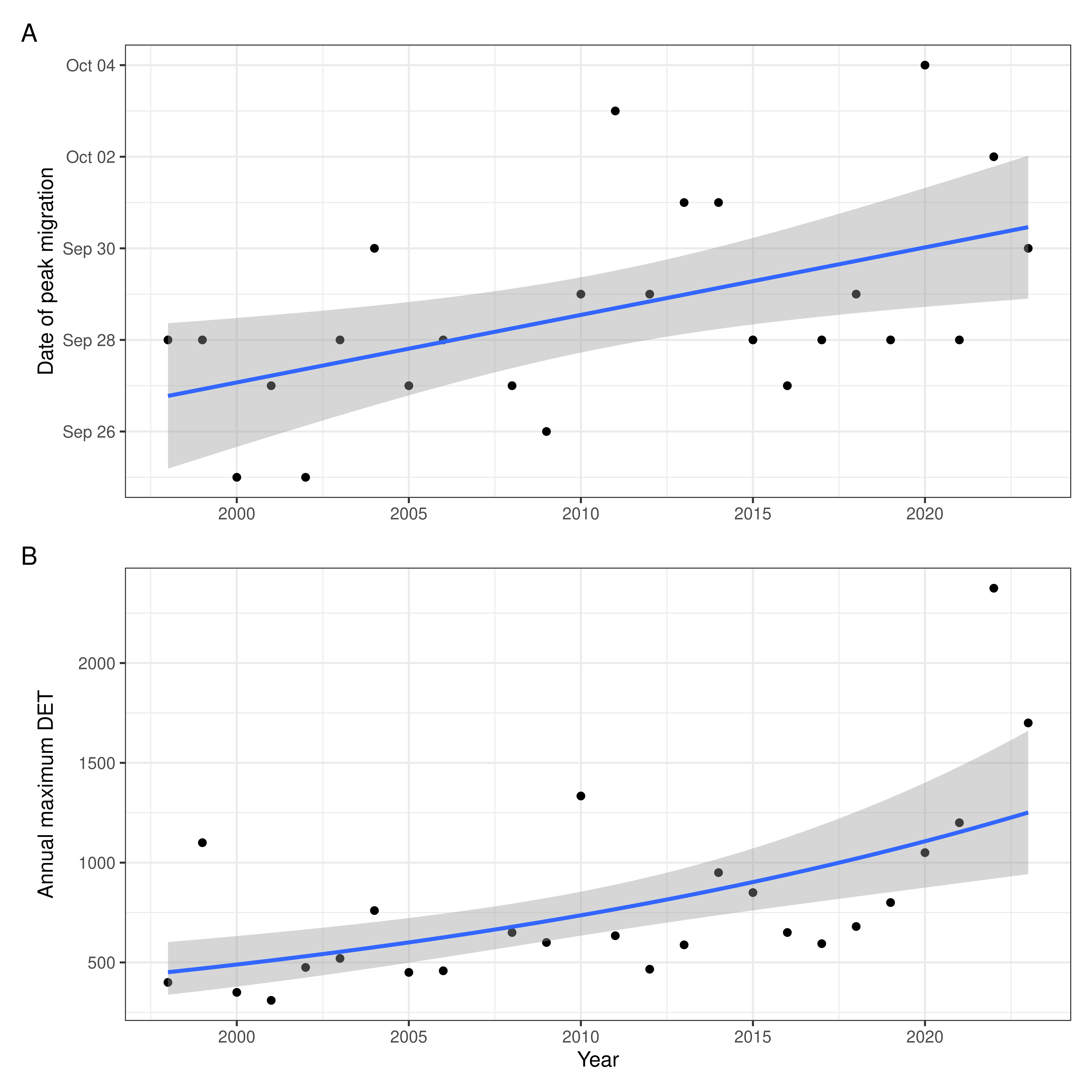
1251/451[1] 2.773836Big version
With two y-axis options for panel B
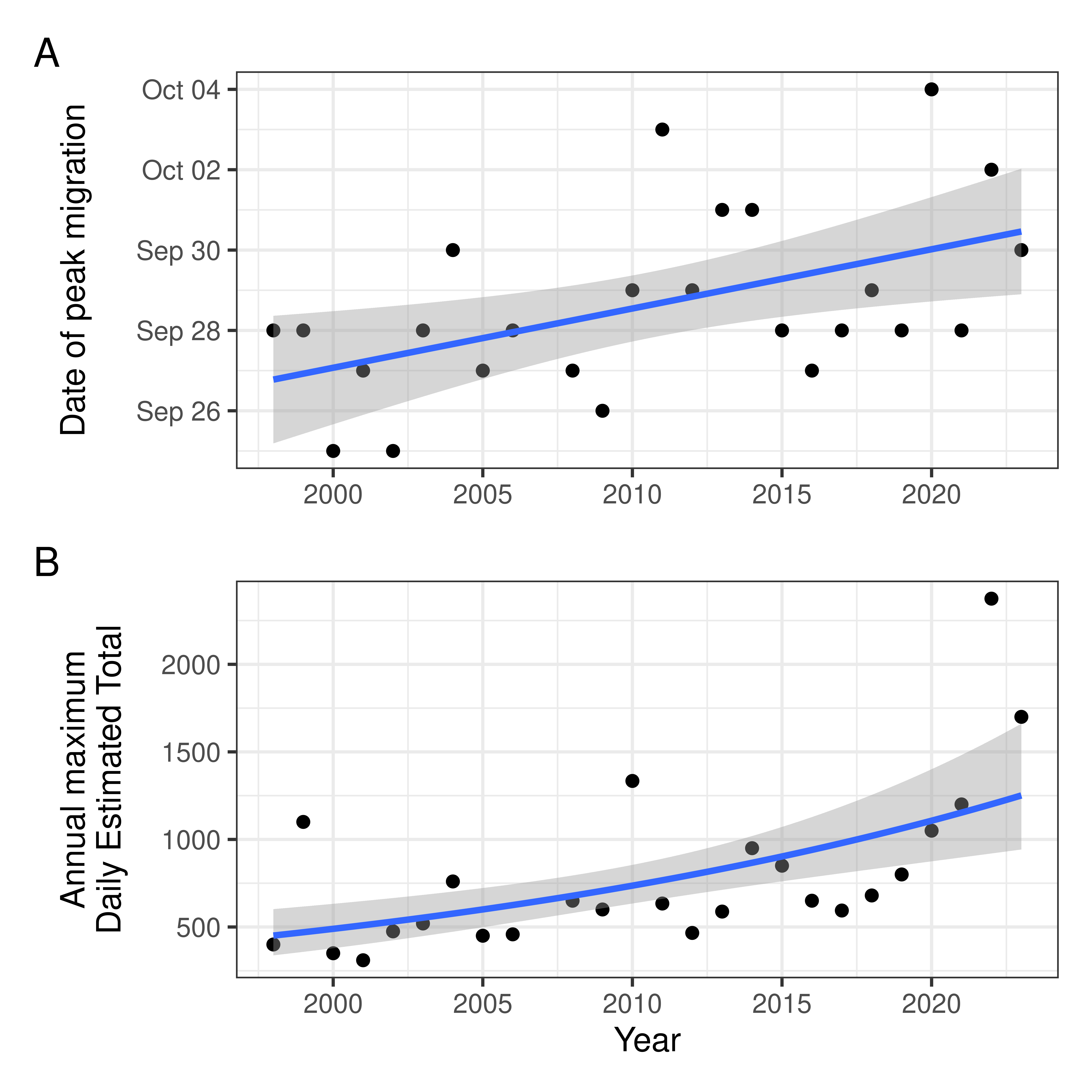
Small version
With two y-axis options for panel B
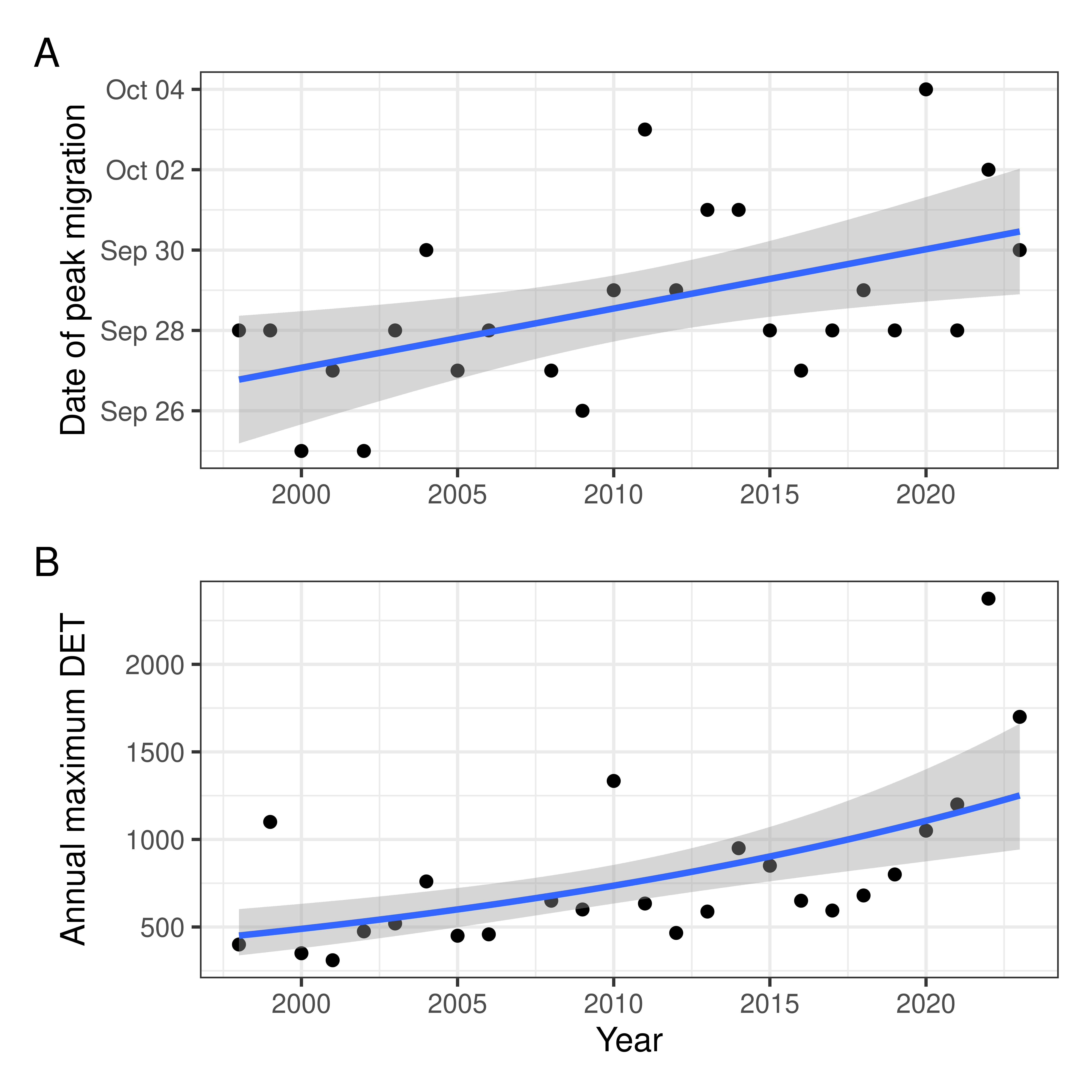
Supplemental comparison of with/without 2022
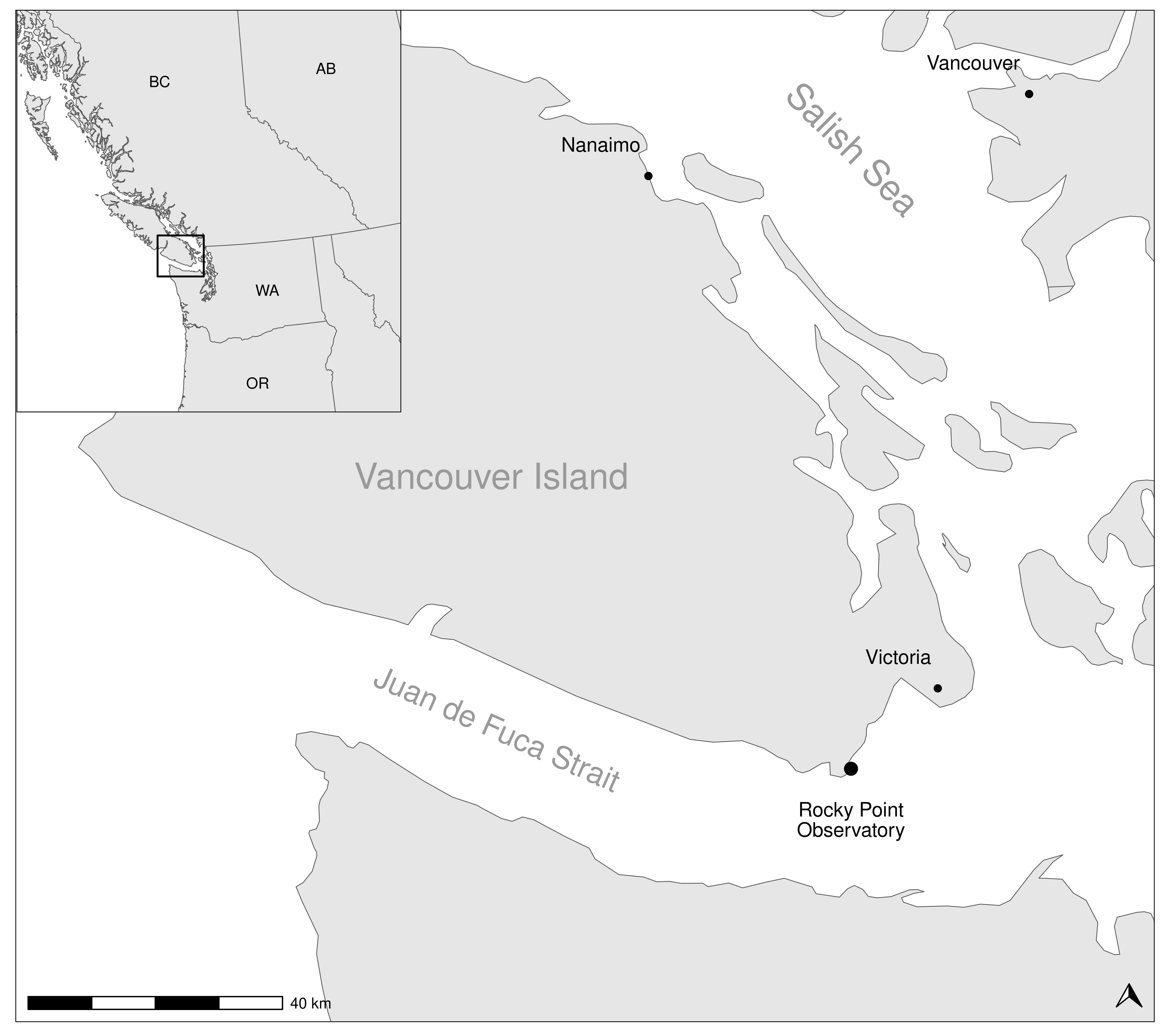
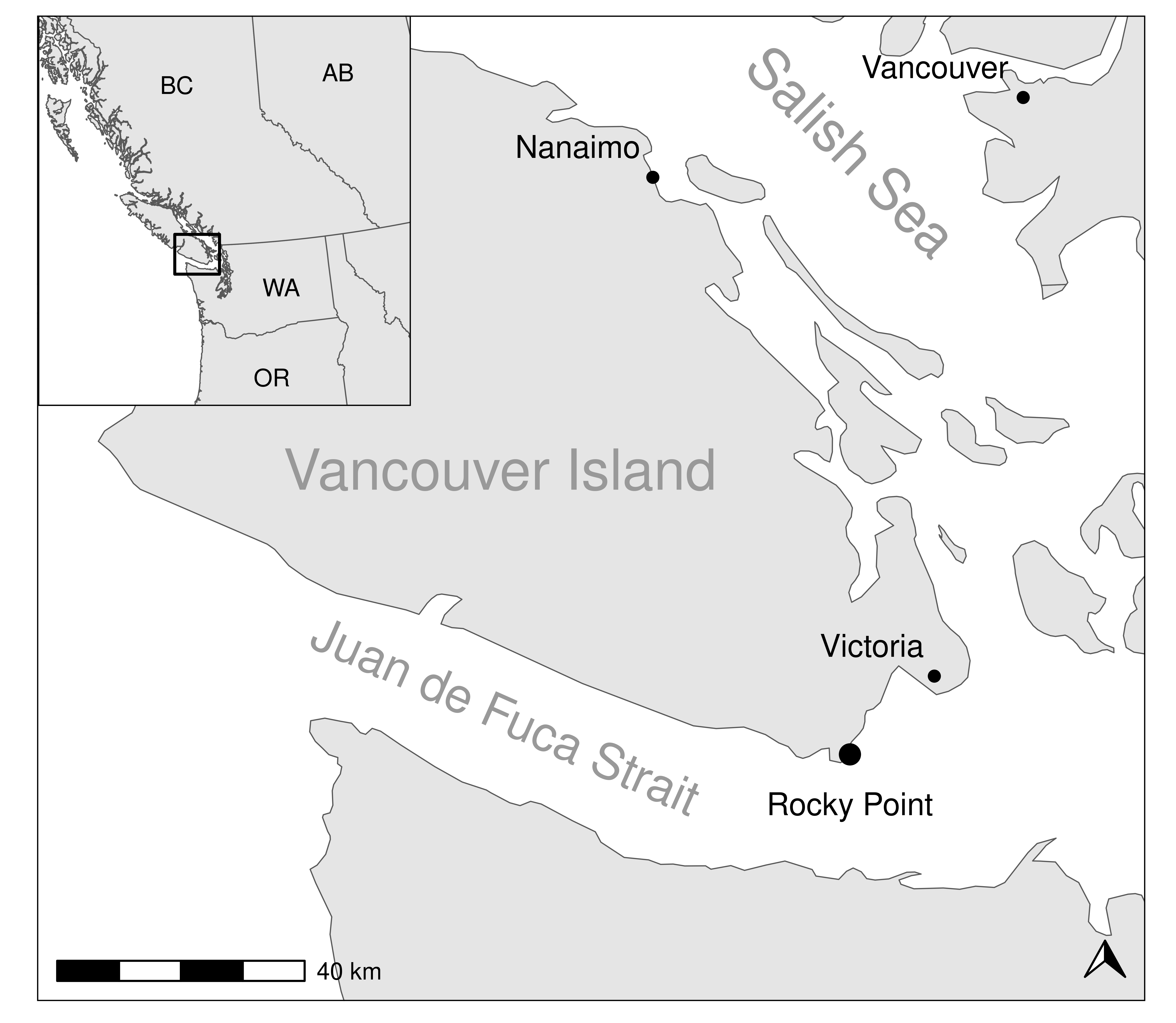
Map
Code
library(bcmaps)
library(ggrepel)
ne_download(type = "populated_places", scale = "large", load = FALSE)[1] "ne_10m_populated_places"Code
cities <- bcmaps::bc_cities(ask = FALSE) |>
select(NAME) |>
filter(NAME %in% c("Victoria", "Vancouver", "Nanaimo"))bc_cities was updated on 2024-07-10Code
cities <- ne_load(file_name = "ne_10m_populated_places") |>
filter(NAME %in% c("Seattle", "Portland"), ADM1NAME %in% c("Oregon", "Washington")) |>
select(NAME) |>
st_transform(st_crs(cities)) |>
bind_rows(cities) |>
rename(name = NAME)Reading layer `ne_10m_populated_places' from data source
`/home/steffi/R_tmpdir/RtmpgJCSAG/ne_10m_populated_places.shp'
using driver `ESRI Shapefile'
Simple feature collection with 7342 features and 137 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -179.59 ymin: -90 xmax: 179.3833 ymax: 82.48332
Geodetic CRS: WGS 84Code
stn <- c(-123.55082035835214, 48.31773308537152) |>
st_point() |>
st_sfc(crs = 4326) |>
st_sf(name = "Rocky Point") |>
st_transform(st_crs(cities))
land <- data.frame(lon = c(-124.3, -124.3, -123.5),
lat = c(48.39, 48.75, 49.2),
name = c("Juan de Fuca Strait", "Vancouver Island", "Salish Sea")) |>
st_as_sf(coords = c("lon", "lat"), crs = 4326)
area <- ne_states(country = c("Canada", "United States of America")) |>
#st_crop(st_bbox(c(xmin = -140, xmax = -112, ymin = 41, ymax = 60))) |>
#filter(name %in% c("British Columbia", "Alberta", "Washington", "Oregon", "Idaho", "Montana")) |>
mutate(
name = if_else(name %in% c("British Columbia", "Washington", "Oregon"), toupper(name), NA),
name = str_replace(name, " ", "\n"),
postal = if_else(postal %in% c("BC", "WA", "OR", "AB"), postal, NA))
box <- st_polygon(list(rbind(c(1050000, 330000),
c(1050000, 490000),
c(1230000, 490000),
c(1230000, 330000),
c(1050000, 330000)))) |>
st_sfc(crs = 3005)
g0 <- ggplot(data = area, aes(label = name)) +
theme_map() +
theme(panel.border = element_rect(fill = NA),
plot.margin = unit(c(0,0,0,0), units = "mm"),
panel.spacing = unit(0, units = "mm")) +
scale_x_continuous(expand = c(0,0)) +
scale_y_continuous(expand = c(0,0)) +
geom_sf()
g <- g0 +
geom_sf(data = stn, size = 3) +
geom_sf_text(data = stn, lineheight = 0.85, nudge_y = -8000) +
geom_sf(data = cities) +
geom_sf_text(data = cities, hjust = 1.1, nudge_y = 5000) +
geom_sf_text(data = land, angle = c(-24, 0, -45), colour = "grey60", size = c(6, 7, 7)) +
annotation_scale(location = "bl") +
annotation_north_arrow(
location = "bl", height = unit(0.5, "cm"), width = unit(0.5, "cm"),
style = north_arrow_orienteering(text_size = -Inf), pad_y = unit(0.75, "cm")) +
coord_sf(crs = 3005, xlim = c(1050000, 1230000), ylim = c(330000, 490000))
g_inset <- g0 +
theme(panel.background = element_rect(fill = "white")) +
geom_sf_text(aes(label = postal), colour = "black", size = 3) +
geom_sf(data = box, fill = NA, linewidth = 0.5, colour = "black", inherit.aes = FALSE) +
coord_sf(crs = 3005, xlim = c(500000, 2000000), ylim = c(-200000, 1370000))Big
Small
Supplemental Figure
Code
pred <- mutate(pred, ci95_upper = count + se * 1.96, ci95_lower = count - se * 1.96)
res <- filter(v, year == 1999) |>
mutate(xmin = 204,
xmax = 240, y = 0.25 * mig_raw_max,
xmid = (240-204)/2 + 204,
y = 0.25 * mig_raw_max,
height = 0.03 * mig_raw_max, label = "Resident period")
g0 <- ggplot() +
theme_void() +
annotate(geom = "text", label = c("Migration\nstart", "Peak\nstart", "Peak", "Peak\nend", "Migration\nend"),
y = -0.25, x = c(0, 0.7, 1.3, 1.6, 2), size = 3, lineheight = 0.85) +
annotate(geom = "text", label = c("5%", "25%", "50%", "75%", "95%"), y = 0.25, x = c(0, 0.7, 1.3, 1.6, 2), size = 3) +
annotate(geom = "segment", y = 0, x = 0, xend = 2, arrow = arrow(angle = 90, ends = "both", length = unit(1, "mm"))) +
annotate(geom = "segment", y = 0, x = 0.7, xend = 1.6, linewidth = 4) +
annotate(geom = "segment", x = 1.29, xend = 1.30, y = 0, linewidth = 8) +
ylim(c(-0.5, 0.5)) +
xlim(c(-0.1, 2.1))
g1 <- ggplot(data = pred, mapping = aes(x = doy, y = count)) +
theme_bw() +
# GAM
geom_ribbon(aes(ymin = ci95_lower, ymax = ci95_upper), fill = "grey50", alpha = 0.5) +
geom_line() +
# Raw points
geom_point(data = raw, na.rm = TRUE, size = 0.5) +
# Metrics
geom_errorbarh(data = res, aes(xmin = xmin, xmax = xmax, y = y, height = height),
colour = "grey70", inherit.aes = FALSE) +
geom_text(data = res, aes(x = xmid, y = y, label = label), vjust = -0.5, inherit.aes = FALSE,
size = 3, colour = "grey30") +
geom_segment(data = v, aes(x = p50_doy - 0.5, xend = p50_doy + 0.5,
y = -(0.07 * mig_raw_max)),
linewidth = 4, inherit.aes = FALSE) +
geom_errorbarh(data = v, aes(y = -(0.07 * mig_raw_max),
xmin = mig_start_doy,
xmax = mig_end_doy,
height = 0.07 * mig_raw_max), inherit.aes = FALSE) +
geom_segment(data = v, aes(y = -(0.07 * mig_raw_max), x = peak_start_doy, xend = peak_end_doy), linewidth = 2, inherit.aes = FALSE) +
scale_x_continuous(name = "Date", limits = c(203, 295),
labels = \(x) format(as_date(x) - days(1), "%b %d"),
n.breaks = 7) +
labs(y = "Daily Estimated Total") +
facet_wrap(~ year, scales = "free_y", ncol = 4)Big
Small
Supplemental Table
Presented here for the record, but use exported XLSX version for submission.
Code
t <- supp |>
filter(!str_detect(term, "(I|i)ntercept"),
!str_detect(model, "(max_doy)|(pop_max)")) |>
mutate(across(where(is.numeric), \(x) round(x, 3))) |>
mutate(p_value = if_else(p_value <= 0.05,
paste0("**", format(p_value, nsmall = 3), "**"),
format(p_value, nsmall = 3)),
value = paste0(format(estimate, nsmall = 3), " (P = ", p_value, ")"),
value = str_trim(value)) |>
select(Model = model, value, type) |>
mutate(type = str_replace(type, "_", " "),
type = str_to_title(type)) |>
pivot_wider(names_from = "type", values_from = "value")
gt(t) |>
gt_theme() |>
fmt_markdown() |>
fmt_number(columns = -1, decimals = 3)| Model | Original | Gaps Removed | End Removed | All Removed |
|---|---|---|---|---|
| mig_start_doy ~ year | 0.206 (P = 0.011) | 0.204 (P = 0.011) | 0.198 (P = 0.026) | 0.196 (P = 0.029) |
| peak_start_doy ~ year | 0.178 (P = 0.003) | 0.176 (P = 0.003) | 0.178 (P = 0.011) | 0.176 (P = 0.010) |
| p50_doy ~ year | 0.164 (P = 0.008) | 0.161 (P = 0.004) | 0.167 (P = 0.023) | 0.165 (P = 0.015) |
| peak_end_doy ~ year | 0.156 (P = 0.013) | 0.151 (P = 0.008) | 0.147 (P = 0.048) | 0.143 (P = 0.036) |
| mig_end_doy ~ year | 0.119 (P = 0.171) | 0.115 (P = 0.192) | 0.077 (P = 0.445) | 0.073 (P = 0.480) |
| mig_dur_days ~ year | -0.087 (P = 0.373) | -0.089 (P = 0.388) | -0.121 (P = 0.297) | -0.123 (P = 0.314) |
| peak_dur_days ~ year | -0.022 (P = 0.563) | -0.024 (P = 0.531) | -0.031 (P = 0.485) | -0.034 (P = 0.457) |
| mig_raw_max ~ year | 0.041 (P = 0.000) | 0.042 (P = 0.000) | 0.033 (P = 0.005) | 0.034 (P = 0.005) |
Code
t <- supp |>
filter(!str_detect(term, "(I|i)ntercept")) |>
mutate(across(where(is.numeric), \(x) round(x, 3))) |>
select(model, type, estimate, p_value) |>
pivot_wider(names_from = "type", values_from = c("estimate", "p_value"),
names_glue = "{type}_{.value}") |>
select(model, contains("original"), contains("gaps"), contains("end"), contains("all")) |>
rename("Model" = "model",
"Original" = "original_estimate",
"Removed Gaps" = "gaps_removed_estimate",
"Removed Ends " = "end_removed_estimate",
"Removed All" = "all_removed_estimate")
wb <- createWorkbook()
addWorksheet(wb, "Supplemental Table 1")
writeData(wb, 1, t)
bold <- createStyle(textDecoration = "bold")
pval <- createStyle(halign = "left", numFmt = "(P = 0.000)")
est <- createStyle(halign = "right", numFmt = "0.000")
for(c in c(3, 5, 7, 9)) {
conditionalFormatting(wb, 1, cols = c, rows = 1:15, rule = "<=0.05", style = bold)
addStyle(wb, 1, cols = c, rows = 1:15, style = pval, stack = TRUE)
}
for(c in c(2, 4, 6, 8)) {
addStyle(wb, 1, cols = c, rows = 1:15, style = est, stack = TRUE)
}
mergeCells(wb, 1, cols = 2:3, rows = 1)
mergeCells(wb, 1, cols = 4:5, rows = 1)
mergeCells(wb, 1, cols = 6:7, rows = 1)
mergeCells(wb, 1, cols = 8:9, rows = 1)
addStyle(wb, 1, cols = 1:9, rows = 1,
style = createStyle(halign = "center", textDecoration = "bold"),
stack = TRUE)
setColWidths(wb, 1, cols = 1:9, widths = 10)
setColWidths(wb, 1, cols = 1, widths = 20)
setRowHeights(wb, 1, rows = nrow(t) + 3, heights = 100)
writeData(
wb, 1, startCol = 1, startRow = nrow(t) + 3,
x = paste0(
"Table. Results [Estimate of Year (P-value)] for different models when removing selective years. \n",
"'Original' represents the results for the original model;\n",
"'Removed Gaps' represents the results for these models when removing years with large gaps around the migration peaks (2011, 2013);\n",
"'Removed Ends' represents the results for these models when removing years where it looks like sampling ended earlier than the end of migration (2001, 2006, 2023);\n",
"'Removed All' represents the results for these models when removing all questionable years (2001, 2006, 2011, 2013, 2023)\n",
"Bold P values are significant at P <= 0.05."
))
saveWorkbook(wb, "Data/table_supp.xlsx", overwrite = TRUE)